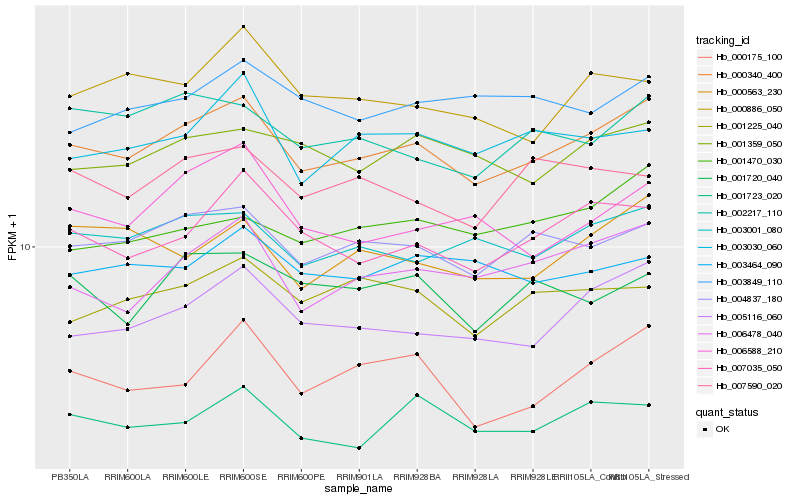
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000340_400 |
0.0 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 2 |
Hb_004837_180 |
0.0563763873 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_005116_060 |
0.0566869657 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003001_080 |
0.0612904479 |
- |
- |
PREDICTED: uncharacterized protein LOC105644363 [Jatropha curcas] |
| 5 |
Hb_006478_040 |
0.063344183 |
- |
- |
PREDICTED: methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial [Jatropha curcas] |
| 6 |
Hb_007035_050 |
0.0653396427 |
- |
- |
PREDICTED: cleavage stimulating factor 64 [Jatropha curcas] |
| 7 |
Hb_001359_050 |
0.0714723541 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000563_230 |
0.0720809275 |
- |
- |
phospholipase d zeta, putative [Ricinus communis] |
| 9 |
Hb_002217_110 |
0.0731558265 |
- |
- |
PREDICTED: craniofacial development protein 1 [Jatropha curcas] |
| 10 |
Hb_001225_040 |
0.0747372107 |
- |
- |
PREDICTED: meiosis-specific nuclear structural protein 1 isoform X5 [Jatropha curcas] |
| 11 |
Hb_003030_060 |
0.0752369555 |
- |
- |
PREDICTED: regulation of nuclear pre-mRNA domain-containing protein 2-like [Jatropha curcas] |
| 12 |
Hb_006588_210 |
0.0753752024 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase Hakai [Jatropha curcas] |
| 13 |
Hb_001723_020 |
0.0760940294 |
- |
- |
PREDICTED: nuclear export mediator factor Nemf [Jatropha curcas] |
| 14 |
Hb_001470_030 |
0.0771465922 |
- |
- |
PREDICTED: MORC family CW-type zinc finger protein 3 isoform X2 [Jatropha curcas] |
| 15 |
Hb_007590_020 |
0.0776695822 |
- |
- |
PREDICTED: protein DAMAGED DNA-BINDING 2 [Jatropha curcas] |
| 16 |
Hb_003464_090 |
0.0779944482 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 17 |
Hb_000886_050 |
0.0782486953 |
- |
- |
hypothetical protein POPTR_0003s01110g [Populus trichocarpa] |
| 18 |
Hb_001720_040 |
0.07912622 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000175_100 |
0.0794413095 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 20 |
Hb_003849_110 |
0.0808003409 |
- |
- |
hypothetical protein JCGZ_17842 [Jatropha curcas] |