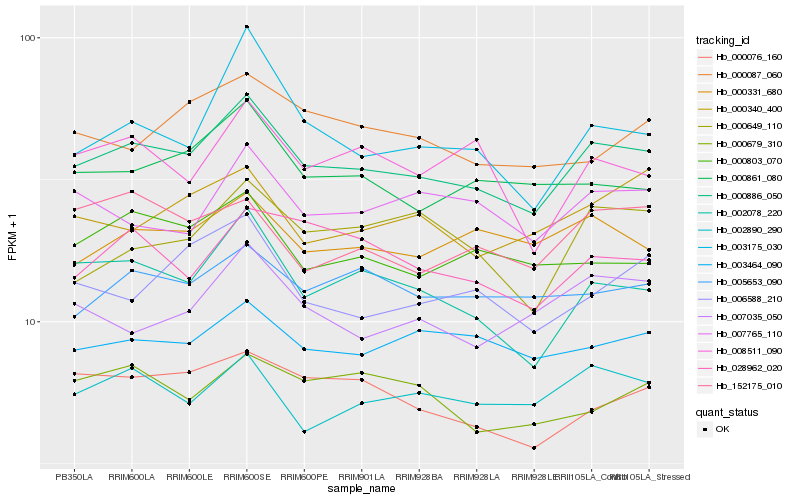
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000886_050 |
0.0 |
- |
- |
hypothetical protein POPTR_0003s01110g [Populus trichocarpa] |
| 2 |
Hb_002078_220 |
0.0596304618 |
- |
- |
PREDICTED: CTD small phosphatase-like protein 2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000861_080 |
0.0697910369 |
- |
- |
PREDICTED: protein ELC-like [Jatropha curcas] |
| 4 |
Hb_002890_290 |
0.0704066928 |
- |
- |
PREDICTED: outer envelope protein 64, mitochondrial [Jatropha curcas] |
| 5 |
Hb_003464_090 |
0.0706141063 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 6 |
Hb_000076_160 |
0.0715533946 |
- |
- |
PREDICTED: AP-5 complex subunit zeta-1 [Jatropha curcas] |
| 7 |
Hb_005653_090 |
0.07160514 |
- |
- |
PREDICTED: protein HIRA isoform X1 [Jatropha curcas] |
| 8 |
Hb_000331_680 |
0.0719970735 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 9 |
Hb_000803_070 |
0.0721369331 |
- |
- |
PREDICTED: protein TRAUCO [Jatropha curcas] |
| 10 |
Hb_028962_020 |
0.0729566442 |
- |
- |
hypothetical protein POPTR_0016s00660g [Populus trichocarpa] |
| 11 |
Hb_003175_030 |
0.0738396368 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 12 |
Hb_007765_110 |
0.0746671648 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 40-like [Jatropha curcas] |
| 13 |
Hb_007035_050 |
0.0772293306 |
- |
- |
PREDICTED: cleavage stimulating factor 64 [Jatropha curcas] |
| 14 |
Hb_152175_010 |
0.0779500278 |
- |
- |
Protein grpE, putative [Ricinus communis] |
| 15 |
Hb_006588_210 |
0.0780531827 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase Hakai [Jatropha curcas] |
| 16 |
Hb_000087_060 |
0.0781105996 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 17 |
Hb_000340_400 |
0.0782486953 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 18 |
Hb_008511_090 |
0.0785324551 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like [Jatropha curcas] |
| 19 |
Hb_000679_310 |
0.0789686482 |
- |
- |
PREDICTED: putative UDP-sugar transporter DDB_G0278631 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000649_110 |
0.0797534149 |
- |
- |
PREDICTED: protein VAC14 homolog [Jatropha curcas] |