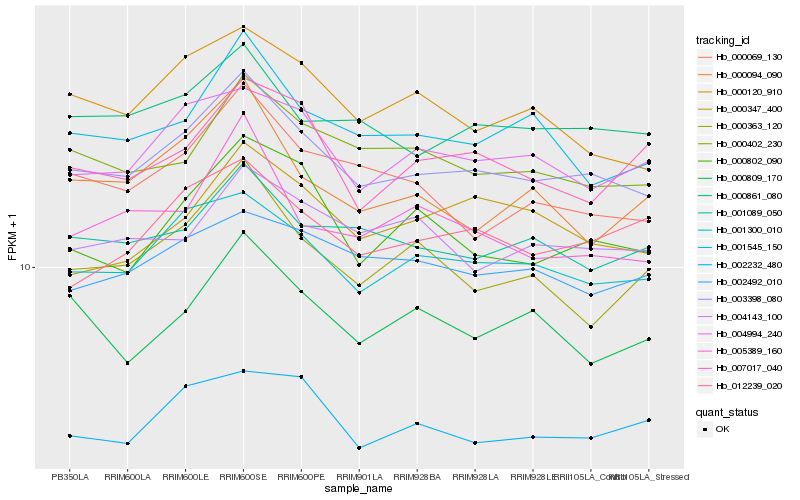
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003398_080 |
0.0 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 2 |
Hb_001300_010 |
0.0514500337 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 3 |
Hb_000861_080 |
0.0620891195 |
- |
- |
PREDICTED: protein ELC-like [Jatropha curcas] |
| 4 |
Hb_000402_230 |
0.0632230911 |
- |
- |
PREDICTED: sorting nexin 2A-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_004994_240 |
0.0647365765 |
- |
- |
hypothetical protein PRUPE_ppa004633mg [Prunus persica] |
| 6 |
Hb_001545_150 |
0.0649630623 |
transcription factor |
TF Family: NAC |
PREDICTED: uncharacterized protein LOC105635527 [Jatropha curcas] |
| 7 |
Hb_000120_910 |
0.0663853617 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 8 |
Hb_001089_050 |
0.0665680885 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 4 [Jatropha curcas] |
| 9 |
Hb_000069_130 |
0.0687328614 |
- |
- |
PREDICTED: uncharacterized protein LOC105642829 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000802_090 |
0.0716165043 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 11 |
Hb_002492_010 |
0.0732544401 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 12 |
Hb_005389_160 |
0.0737958659 |
- |
- |
PREDICTED: casein kinase I [Jatropha curcas] |
| 13 |
Hb_004143_100 |
0.0742359723 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_012239_020 |
0.0754690096 |
- |
- |
PREDICTED: protein VAC14 homolog [Jatropha curcas] |
| 15 |
Hb_000363_120 |
0.075824385 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |
| 16 |
Hb_002232_480 |
0.0759919568 |
- |
- |
PREDICTED: telomerase reverse transcriptase [Jatropha curcas] |
| 17 |
Hb_000094_090 |
0.0782314266 |
- |
- |
PREDICTED: DUF21 domain-containing protein At1g47330 isoform X1 [Jatropha curcas] |
| 18 |
Hb_007017_040 |
0.0783325771 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit [Jatropha curcas] |
| 19 |
Hb_000809_170 |
0.0793142192 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein 1 homolog isoform X1 [Jatropha curcas] |
| 20 |
Hb_000347_400 |
0.0793840901 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |