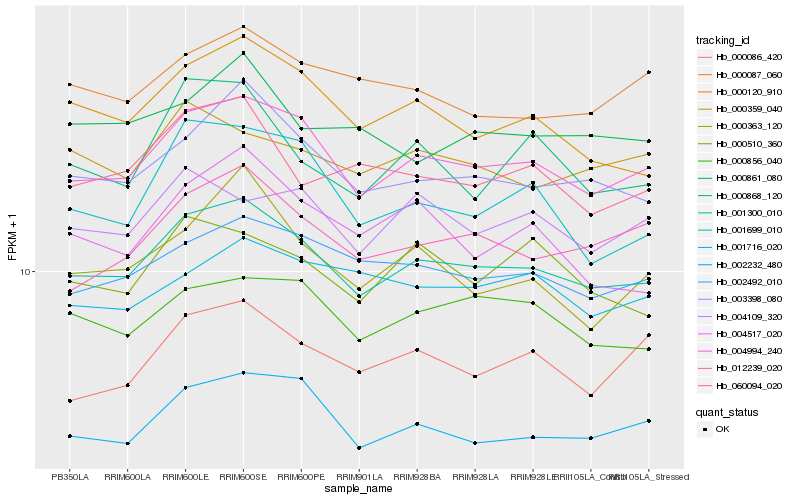
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001300_010 |
0.0 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 2 |
Hb_004994_240 |
0.0287258522 |
- |
- |
hypothetical protein PRUPE_ppa004633mg [Prunus persica] |
| 3 |
Hb_003398_080 |
0.0514500337 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 4 |
Hb_002492_010 |
0.0606669361 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 5 |
Hb_000120_910 |
0.0613032428 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 6 |
Hb_002232_480 |
0.0631005285 |
- |
- |
PREDICTED: telomerase reverse transcriptase [Jatropha curcas] |
| 7 |
Hb_000856_040 |
0.0644073898 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105640470 [Jatropha curcas] |
| 8 |
Hb_012239_020 |
0.0648261538 |
- |
- |
PREDICTED: protein VAC14 homolog [Jatropha curcas] |
| 9 |
Hb_060094_020 |
0.0689433412 |
desease resistance |
Gene Name: ATP_bind_1 |
xpa-binding protein, putative [Ricinus communis] |
| 10 |
Hb_001699_010 |
0.0734286648 |
- |
- |
drought-inducible protein [Manihot esculenta] |
| 11 |
Hb_000510_360 |
0.0737685206 |
transcription factor |
TF Family: GNAT |
PREDICTED: histone acetyltransferase GCN5 [Jatropha curcas] |
| 12 |
Hb_000087_060 |
0.0746454527 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 13 |
Hb_000868_120 |
0.0750367961 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 2 [Jatropha curcas] |
| 14 |
Hb_004109_320 |
0.0751815669 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 52 A [Jatropha curcas] |
| 15 |
Hb_000861_080 |
0.0759833499 |
- |
- |
PREDICTED: protein ELC-like [Jatropha curcas] |
| 16 |
Hb_000086_420 |
0.076046756 |
- |
- |
PREDICTED: phospholipid--sterol O-acyltransferase [Jatropha curcas] |
| 17 |
Hb_000359_040 |
0.0766881681 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_004517_020 |
0.0768484275 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2B isoform X1 [Jatropha curcas] |
| 19 |
Hb_000363_120 |
0.0776081355 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |
| 20 |
Hb_001716_020 |
0.0779164356 |
- |
- |
hydrolase, putative [Ricinus communis] |