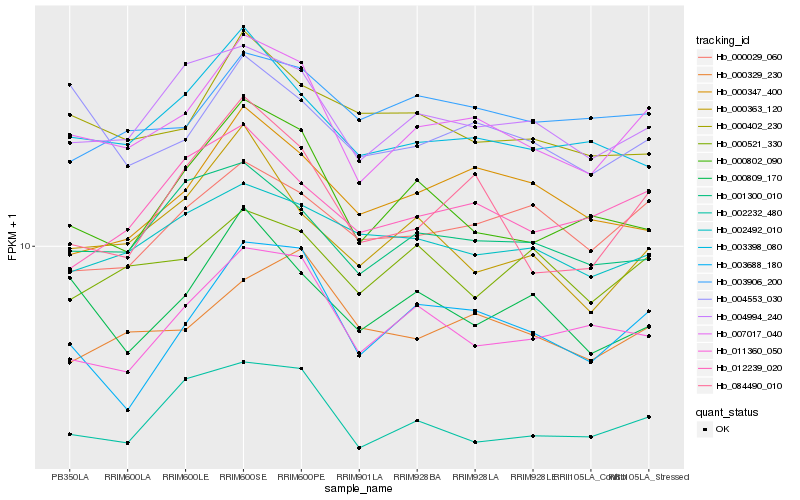
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007017_040 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit [Jatropha curcas] |
| 2 |
Hb_004994_240 |
0.0657095583 |
- |
- |
hypothetical protein PRUPE_ppa004633mg [Prunus persica] |
| 3 |
Hb_002232_480 |
0.0689717082 |
- |
- |
PREDICTED: telomerase reverse transcriptase [Jatropha curcas] |
| 4 |
Hb_084490_010 |
0.0757365393 |
- |
- |
PREDICTED: protein root UVB sensitive 6 isoform X2 [Jatropha curcas] |
| 5 |
Hb_003688_180 |
0.076184014 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 6 |
Hb_003398_080 |
0.0783325771 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 7 |
Hb_011360_050 |
0.0787824829 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 8, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001300_010 |
0.0793607036 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 9 |
Hb_000802_090 |
0.0804514032 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 10 |
Hb_004553_030 |
0.0808783982 |
- |
- |
PREDICTED: uncharacterized protein LOC105629320 [Jatropha curcas] |
| 11 |
Hb_000029_060 |
0.0818345475 |
- |
- |
PREDICTED: mRNA-decapping enzyme-like protein [Jatropha curcas] |
| 12 |
Hb_000329_230 |
0.0839775697 |
- |
- |
PREDICTED: uncharacterized protein LOC105643152 [Jatropha curcas] |
| 13 |
Hb_000521_330 |
0.0845075647 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_012239_020 |
0.084558826 |
- |
- |
PREDICTED: protein VAC14 homolog [Jatropha curcas] |
| 15 |
Hb_003906_200 |
0.0855823176 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48 [Jatropha curcas] |
| 16 |
Hb_002492_010 |
0.0863530272 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 17 |
Hb_000402_230 |
0.0865848346 |
- |
- |
PREDICTED: sorting nexin 2A-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_000809_170 |
0.0869015846 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein 1 homolog isoform X1 [Jatropha curcas] |
| 19 |
Hb_000363_120 |
0.0887013091 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 20 [Jatropha curcas] |
| 20 |
Hb_000347_400 |
0.0888544073 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |