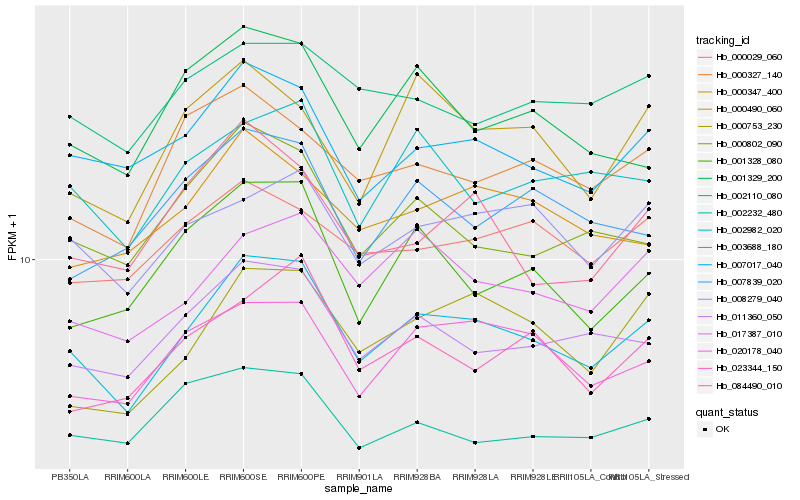
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003688_180 |
0.0 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 2 |
Hb_011360_050 |
0.0745829033 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 8, chloroplastic [Jatropha curcas] |
| 3 |
Hb_007017_040 |
0.076184014 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit [Jatropha curcas] |
| 4 |
Hb_020178_040 |
0.0809479009 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_017387_010 |
0.0810534342 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At4g35230 [Jatropha curcas] |
| 6 |
Hb_000753_230 |
0.0846825503 |
- |
- |
PREDICTED: probable galacturonosyltransferase 11 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002982_020 |
0.0855815837 |
- |
- |
PREDICTED: putative ER lumen protein-retaining receptor C28H8.4 [Jatropha curcas] |
| 8 |
Hb_008279_040 |
0.086911608 |
- |
- |
Rer1 family protein isoform 2 [Theobroma cacao] |
| 9 |
Hb_000802_090 |
0.0882506661 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 10 |
Hb_000347_400 |
0.0933398757 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001328_080 |
0.0934470802 |
- |
- |
PREDICTED: importin subunit alpha-1b [Jatropha curcas] |
| 12 |
Hb_000327_140 |
0.0935588517 |
- |
- |
PREDICTED: probable adenylate kinase 6, chloroplastic [Jatropha curcas] |
| 13 |
Hb_023344_150 |
0.0936593757 |
- |
- |
PREDICTED: uncharacterized protein LOC105644462 [Jatropha curcas] |
| 14 |
Hb_001329_200 |
0.0939343891 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 1 [Jatropha curcas] |
| 15 |
Hb_002110_080 |
0.0960123968 |
- |
- |
FH interacting protein 1 [Theobroma cacao] |
| 16 |
Hb_084490_010 |
0.0970100887 |
- |
- |
PREDICTED: protein root UVB sensitive 6 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000490_060 |
0.097764112 |
- |
- |
PREDICTED: splicing factor 3A subunit 2 [Jatropha curcas] |
| 18 |
Hb_002232_480 |
0.0980221502 |
- |
- |
PREDICTED: telomerase reverse transcriptase [Jatropha curcas] |
| 19 |
Hb_000029_060 |
0.0981964291 |
- |
- |
PREDICTED: mRNA-decapping enzyme-like protein [Jatropha curcas] |
| 20 |
Hb_007839_020 |
0.0987508384 |
- |
- |
conserved hypothetical protein [Ricinus communis] |