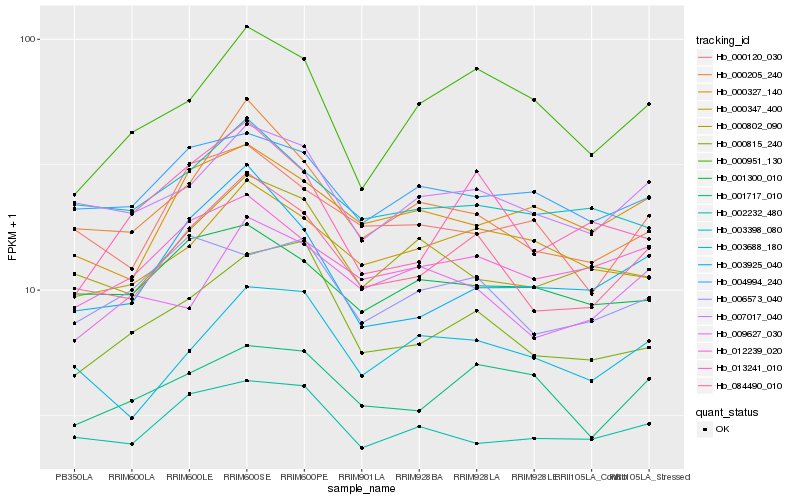
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_084490_010 |
0.0 |
- |
- |
PREDICTED: protein root UVB sensitive 6 isoform X2 [Jatropha curcas] |
| 2 |
Hb_007017_040 |
0.0757365393 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit [Jatropha curcas] |
| 3 |
Hb_000205_240 |
0.0775334172 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 4 |
Hb_012239_020 |
0.0899130218 |
- |
- |
PREDICTED: protein VAC14 homolog [Jatropha curcas] |
| 5 |
Hb_003925_040 |
0.0970074595 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger AN1 and C2H2 domain-containing stress-associated protein 16 [Jatropha curcas] |
| 6 |
Hb_003688_180 |
0.0970100887 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 7 |
Hb_000815_240 |
0.0973339312 |
- |
- |
PREDICTED: galactoside 2-alpha-L-fucosyltransferase-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_002232_480 |
0.0975801018 |
- |
- |
PREDICTED: telomerase reverse transcriptase [Jatropha curcas] |
| 9 |
Hb_000802_090 |
0.0980169543 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 10 |
Hb_000120_030 |
0.0989138006 |
- |
- |
PREDICTED: uncharacterized protein LOC105630385 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003398_080 |
0.1000445936 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 12 |
Hb_004994_240 |
0.100821272 |
- |
- |
hypothetical protein PRUPE_ppa004633mg [Prunus persica] |
| 13 |
Hb_000327_140 |
0.1051078688 |
- |
- |
PREDICTED: probable adenylate kinase 6, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001717_010 |
0.1066103284 |
- |
- |
F-box and wd40 domain protein, putative [Ricinus communis] |
| 15 |
Hb_013241_010 |
0.1067253337 |
- |
- |
PREDICTED: small G protein signaling modulator 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000347_400 |
0.1069645274 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000951_130 |
0.107564777 |
- |
- |
PREDICTED: transcription factor GTE2-like [Jatropha curcas] |
| 18 |
Hb_001300_010 |
0.1077624247 |
transcription factor |
TF Family: C3H |
nucleic acid binding protein, putative [Ricinus communis] |
| 19 |
Hb_009627_030 |
0.1079802738 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g23880-like [Jatropha curcas] |
| 20 |
Hb_006573_040 |
0.1084131087 |
- |
- |
DNA binding protein, putative [Ricinus communis] |