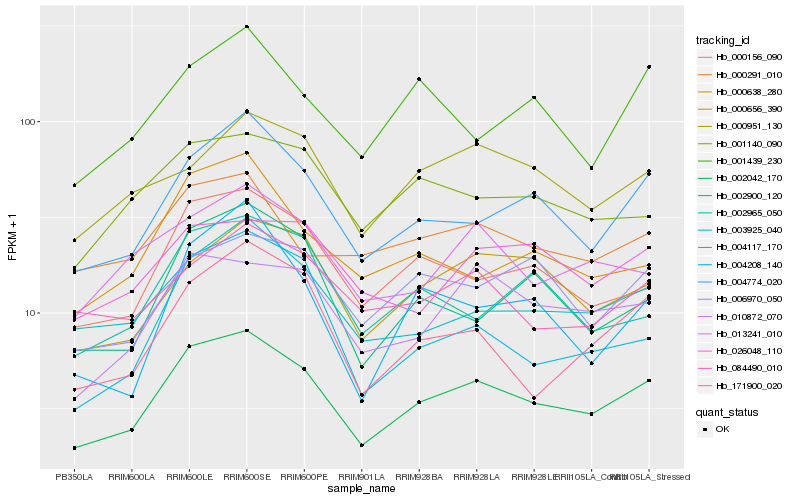
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002042_170 |
0.0 |
- |
- |
RING/U-box superfamily protein [Theobroma cacao] |
| 2 |
Hb_004774_020 |
0.0894068864 |
- |
- |
PREDICTED: PRA1 family protein B4-like [Camelina sativa] |
| 3 |
Hb_000156_090 |
0.0997492105 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS3 isoform X1 [Jatropha curcas] |
| 4 |
Hb_003925_040 |
0.1009764636 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger AN1 and C2H2 domain-containing stress-associated protein 16 [Jatropha curcas] |
| 5 |
Hb_010872_070 |
0.1050296843 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000656_390 |
0.111248758 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |
| 7 |
Hb_171900_020 |
0.1117019543 |
- |
- |
PREDICTED: uncharacterized protein LOC105643207 [Jatropha curcas] |
| 8 |
Hb_002900_120 |
0.1135615489 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 [Jatropha curcas] |
| 9 |
Hb_013241_010 |
0.1192299877 |
- |
- |
PREDICTED: small G protein signaling modulator 2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001140_090 |
0.1198062303 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004117_170 |
0.1212382957 |
- |
- |
PREDICTED: probable receptor-like protein kinase At3g55450 [Jatropha curcas] |
| 12 |
Hb_006970_050 |
0.1221047197 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 13 |
Hb_000951_130 |
0.122750772 |
- |
- |
PREDICTED: transcription factor GTE2-like [Jatropha curcas] |
| 14 |
Hb_000638_280 |
0.123337371 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 15 |
Hb_084490_010 |
0.1237985273 |
- |
- |
PREDICTED: protein root UVB sensitive 6 isoform X2 [Jatropha curcas] |
| 16 |
Hb_004208_140 |
0.1241558105 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g47070 [Jatropha curcas] |
| 17 |
Hb_002965_050 |
0.1249152736 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR2 [Jatropha curcas] |
| 18 |
Hb_026048_110 |
0.1276977573 |
- |
- |
PREDICTED: uncharacterized protein LOC105645733 [Jatropha curcas] |
| 19 |
Hb_000291_010 |
0.1282890851 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 20 |
Hb_001439_230 |
0.1296193402 |
- |
- |
PREDICTED: cystathionine gamma-synthase 1, chloroplastic [Jatropha curcas] |