| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
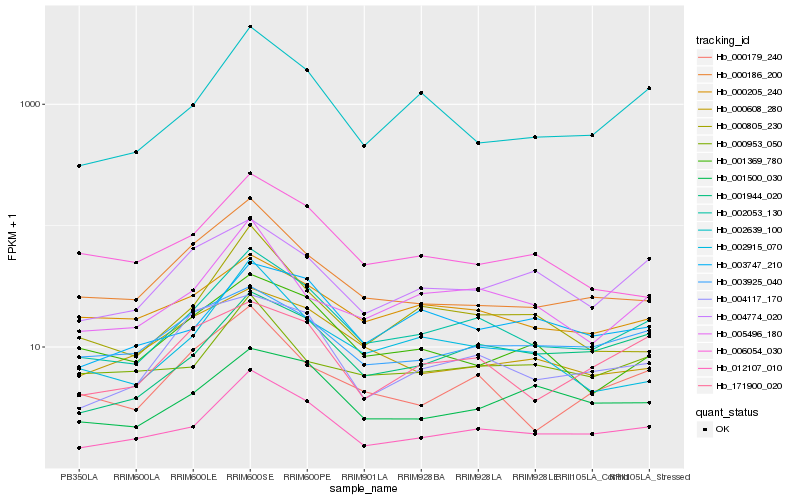
Hb_002053_130 |
0.0 |
- |
- |
UDP-glucose 4-epimerase, putative [Ricinus communis] |
| 2 |
Hb_012107_010 |
0.0760417463 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 3 |
Hb_002639_100 |
0.118774941 |
- |
- |
dehydrin [Hevea brasiliensis] |
| 4 |
Hb_005496_180 |
0.1200914151 |
- |
- |
PREDICTED: U-box domain-containing protein 4-like [Jatropha curcas] |
| 5 |
Hb_001944_020 |
0.1203925712 |
- |
- |
PREDICTED: uncharacterized protein LOC105636535 [Jatropha curcas] |
| 6 |
Hb_000179_240 |
0.1247149066 |
- |
- |
PREDICTED: threonine synthase 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_171900_020 |
0.125923614 |
- |
- |
PREDICTED: uncharacterized protein LOC105643207 [Jatropha curcas] |
| 8 |
Hb_000186_200 |
0.1273756918 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 9 |
Hb_003747_210 |
0.1301609607 |
- |
- |
PREDICTED: uncharacterized protein LOC105630240 [Jatropha curcas] |
| 10 |
Hb_002915_070 |
0.1305293249 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA GLABRA 1 [Jatropha curcas] |
| 11 |
Hb_001369_780 |
0.1310518792 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000205_240 |
0.1317312363 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 13 |
Hb_000805_230 |
0.1339093885 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 14 |
Hb_004774_020 |
0.134304949 |
- |
- |
PREDICTED: PRA1 family protein B4-like [Camelina sativa] |
| 15 |
Hb_003925_040 |
0.1364919194 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger AN1 and C2H2 domain-containing stress-associated protein 16 [Jatropha curcas] |
| 16 |
Hb_001500_030 |
0.1370131403 |
- |
- |
PREDICTED: uncharacterized protein At2g33490 isoform X2 [Jatropha curcas] |
| 17 |
Hb_004117_170 |
0.1372723157 |
- |
- |
PREDICTED: probable receptor-like protein kinase At3g55450 [Jatropha curcas] |
| 18 |
Hb_006054_030 |
0.1383391684 |
- |
- |
PREDICTED: protein GPR107-like [Jatropha curcas] |
| 19 |
Hb_000953_050 |
0.1421779033 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105629451 [Jatropha curcas] |
| 20 |
Hb_000608_280 |
0.1429375907 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |