Hb_002639_100
Information
| Type | - |
|---|---|
| Description | - |
| Location | Contig2639: 91780-93132 |
| Sequence |   |
Annotation
kegg
| ID | cic:CICLE_v10002349mg |
|---|---|
| description | hypothetical protein |
nr
| ID | AJK30589.1 |
|---|---|
| description | dehydrin [Hevea brasiliensis] |
swissprot
| ID | P31168 |
|---|---|
| description | Dehydrin COR47 OS=Arabidopsis thaliana GN=COR47 PE=1 SV=2 |
trembl
| ID | V9M4U6 |
|---|---|
| description | Dehydrin protein OS=Manihot esculenta GN=COR47 PE=2 SV=1 |
Gene Ontology
| ID | GO:0006950 |
|---|---|
| description | dehydrin |
Full-length cDNA clone information
| cDNA+EST (Sanger&Illumina) (ID:Location) |
PASA_asmbl_27563: 91634-93069 , PASA_asmbl_27564: 91907-92866 |
|---|---|
| cDNA (Sanger) (ID:Location) |
006_H06.ab1: 92635-98312 , 007_A02.ab1: 92066-92904 , 010_P16.ab1: 92353-92583 , 011_I11.ab1: 92353-92611 , 013_B04.ab1: 92287-92605 , 017_P06.ab1: 92635-92866 , 018_B17.ab1: 92289-92761 , 018_M06.ab1: 92287-92866 , 023_H14.ab1: 92125-92866 , 027_N11.ab1: 92001-92822 , 034_I13.ab1: 92355-92866 , 035_H22.ab1: 92287-92866 , 036_M04.ab1: 91992-92866 , 039_O08.ab1: 92491-92852 , 040_O04.ab1: 92355-92866 , 040_O19.ab1: 92635-98312 , 041_E16.ab1: 91998-92866 , 041_K03.ab1: 92320-92866 , 041_M02.ab1: 92208-92681 , 044_B24.ab1: 92004-92866 , 044_J05.ab1: 92290-92547 , 047_F20.ab1: 92287-92866 , 047_M20.ab1: 92004-92866 , 048_M09.ab1: 92010-92904 , 049_B18.ab1: 92355-92866 , 050_A22.ab1: 92287-92544 |
Similar expressed genes (Top20)
Gene co-expression network
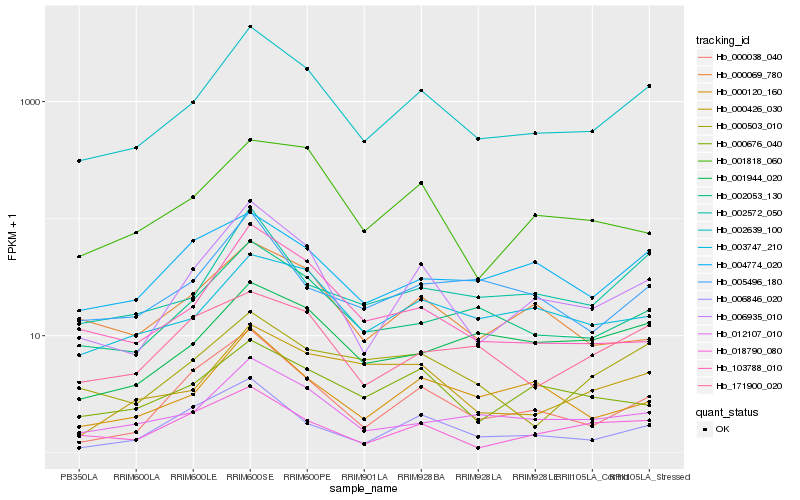
Expression pattern by RNA-Seq analysis
| RRIM600_Latex | RRIM600_Bark | RRIM600_Leaf | RRIM600_Petiole | PB350_Latex | RRIM901_Latex |
|---|---|---|---|---|---|
| 402.472 | 4368.14 | 981.894 | 1904.08 | 310.235 | 453.777 |
| RRII105_Latex_C | RRII105_Latex_S | RRIM928_Latex | RRIM928_Bark | RRIM928_Leaf | |
| 554.704 | 1357.05 | 478.445 | 1242.31 | 534.829 |

