| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
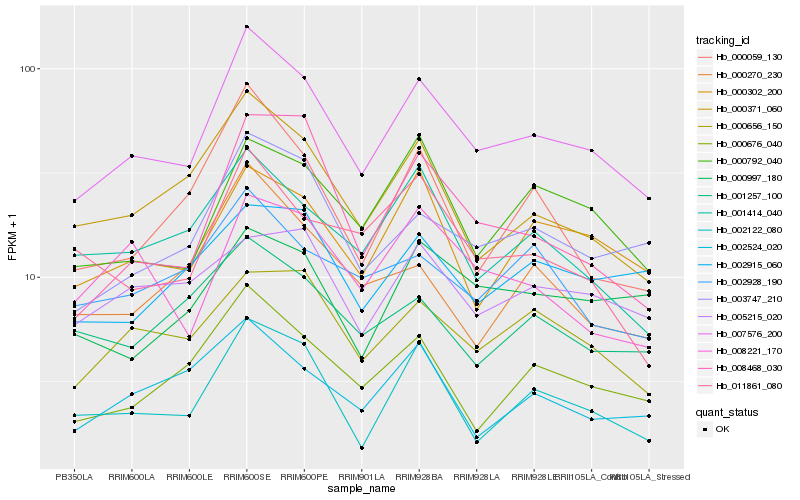
Hb_007576_200 |
0.0 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 2 |
Hb_003747_210 |
0.0990890266 |
- |
- |
PREDICTED: uncharacterized protein LOC105630240 [Jatropha curcas] |
| 3 |
Hb_000371_060 |
0.1051752999 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 4 |
Hb_000676_040 |
0.1114980327 |
- |
- |
hypothetical protein POPTR_0007s13320g [Populus trichocarpa] |
| 5 |
Hb_000656_150 |
0.1131246965 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Jatropha curcas] |
| 6 |
Hb_000302_200 |
0.1133794609 |
- |
- |
ornithine cyclodeaminase, putative [Ricinus communis] |
| 7 |
Hb_002122_080 |
0.113575959 |
- |
- |
- |
| 8 |
Hb_000792_040 |
0.1163240774 |
- |
- |
PREDICTED: steroid 5-alpha-reductase DET2 [Jatropha curcas] |
| 9 |
Hb_001414_040 |
0.1178434819 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_008468_030 |
0.1221348754 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 11 |
Hb_000059_130 |
0.1230127664 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 12 |
Hb_002928_190 |
0.1298022094 |
transcription factor |
TF Family: RWP-RK |
PREDICTED: protein NLP4 [Jatropha curcas] |
| 13 |
Hb_002524_020 |
0.1305014378 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 14 |
Hb_001257_100 |
0.1318690819 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor E2FB isoform X1 [Jatropha curcas] |
| 15 |
Hb_008221_170 |
0.1326815851 |
transcription factor |
TF Family: BES1 |
PREDICTED: BES1/BZR1 homolog protein 4 isoform X1 [Populus euphratica] |
| 16 |
Hb_002915_060 |
0.1329683538 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1-like [Jatropha curcas] |
| 17 |
Hb_011861_080 |
0.1331608142 |
- |
- |
hypothetical protein JCGZ_06088 [Jatropha curcas] |
| 18 |
Hb_000997_180 |
0.1338191031 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_005215_020 |
0.1339790484 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB44 [Jatropha curcas] |
| 20 |
Hb_000270_230 |
0.134081993 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 19 [Jatropha curcas] |