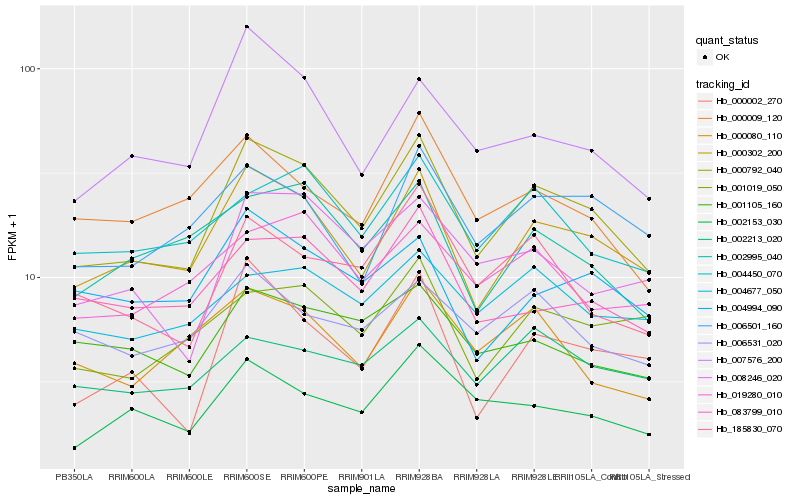
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000792_040 |
0.0 |
- |
- |
PREDICTED: steroid 5-alpha-reductase DET2 [Jatropha curcas] |
| 2 |
Hb_000302_200 |
0.0672751436 |
- |
- |
ornithine cyclodeaminase, putative [Ricinus communis] |
| 3 |
Hb_185830_070 |
0.1133042323 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_004450_070 |
0.1159521623 |
- |
- |
PREDICTED: 2-hydroxyacyl-CoA lyase [Jatropha curcas] |
| 5 |
Hb_007576_200 |
0.1163240774 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 6 |
Hb_006501_160 |
0.1171288346 |
- |
- |
PREDICTED: hexokinase-1-like [Jatropha curcas] |
| 7 |
Hb_008246_020 |
0.1179047829 |
- |
- |
amidophosphoribosyltransferase, putative [Ricinus communis] |
| 8 |
Hb_002153_030 |
0.1183421327 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Theobroma cacao] |
| 9 |
Hb_083799_010 |
0.1197534319 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 10 |
Hb_000002_270 |
0.1207203191 |
- |
- |
hypothetical protein VITISV_043897 [Vitis vinifera] |
| 11 |
Hb_002995_040 |
0.1214639139 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase dyrk1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_019280_010 |
0.1234575756 |
- |
- |
PREDICTED: F-box protein At5g39450 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000009_120 |
0.1237337253 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P isoform X1 [Jatropha curcas] |
| 14 |
Hb_002213_020 |
0.1242615077 |
- |
- |
PREDICTED: uncharacterized protein LOC105630320 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000080_110 |
0.1246489316 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001019_050 |
0.1254176861 |
- |
- |
PREDICTED: importin-13 [Jatropha curcas] |
| 17 |
Hb_006531_020 |
0.12621817 |
- |
- |
PREDICTED: autophagy-related protein 13 [Jatropha curcas] |
| 18 |
Hb_001105_160 |
0.1262216513 |
- |
- |
PREDICTED: protein phosphatase 2C 32 [Jatropha curcas] |
| 19 |
Hb_004994_090 |
0.1267404761 |
- |
- |
PREDICTED: uncharacterized protein LOC105643033 isoform X2 [Jatropha curcas] |
| 20 |
Hb_004677_050 |
0.1269607831 |
- |
- |
PREDICTED: importin beta-like SAD2 isoform X2 [Jatropha curcas] |