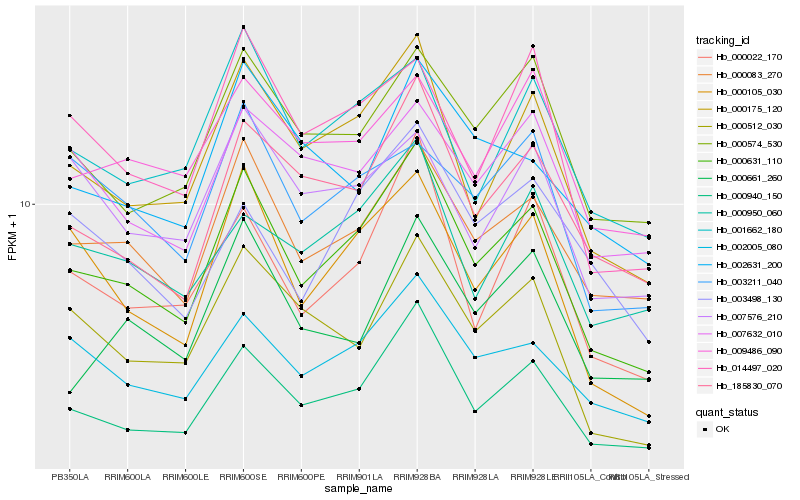
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000631_110 |
0.0 |
- |
- |
PREDICTED: activating signal cointegrator 1 complex subunit 3 [Jatropha curcas] |
| 2 |
Hb_000940_150 |
0.0568521109 |
- |
- |
PREDICTED: uncharacterized protein LOC105643779 [Jatropha curcas] |
| 3 |
Hb_000175_120 |
0.0712992196 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 4 |
Hb_000083_270 |
0.0797507599 |
- |
- |
PREDICTED: putative uncharacterized protein At4g01020, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000105_030 |
0.0889761071 |
- |
- |
fkbp-rapamycin associated protein, putative [Ricinus communis] |
| 6 |
Hb_002005_080 |
0.0904654314 |
- |
- |
PREDICTED: uncharacterized protein LOC105649750 isoform X1 [Jatropha curcas] |
| 7 |
Hb_185830_070 |
0.0918353539 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000022_170 |
0.0986690136 |
- |
- |
PREDICTED: uncharacterized protein LOC105628473 [Jatropha curcas] |
| 9 |
Hb_000574_530 |
0.1003120151 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 10 |
Hb_001662_180 |
0.1048578266 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit 1-like isoform X5 [Citrus sinensis] |
| 11 |
Hb_014497_020 |
0.1068580435 |
- |
- |
PREDICTED: U5 small nuclear ribonucleoprotein 200 kDa helicase [Jatropha curcas] |
| 12 |
Hb_000512_030 |
0.1085648107 |
- |
- |
wall-associated kinase, putative [Ricinus communis] |
| 13 |
Hb_000950_060 |
0.1122782969 |
- |
- |
PREDICTED: CCAAT/enhancer-binding protein zeta [Jatropha curcas] |
| 14 |
Hb_009486_090 |
0.1183251352 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_000661_260 |
0.1196271164 |
- |
- |
PREDICTED: uncharacterized protein LOC105648304 [Jatropha curcas] |
| 16 |
Hb_007632_010 |
0.1203810365 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003498_130 |
0.1209438191 |
- |
- |
PREDICTED: protein ROOT HAIR DEFECTIVE 3 homolog 2-like [Citrus sinensis] |
| 18 |
Hb_003211_040 |
0.1211224187 |
- |
- |
PREDICTED: uncharacterized protein LOC105634664 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002631_200 |
0.1218832972 |
- |
- |
PREDICTED: uncharacterized protein LOC105646171 isoform X2 [Jatropha curcas] |
| 20 |
Hb_007576_210 |
0.1222408826 |
- |
- |
PREDICTED: protein furry homolog [Jatropha curcas] |