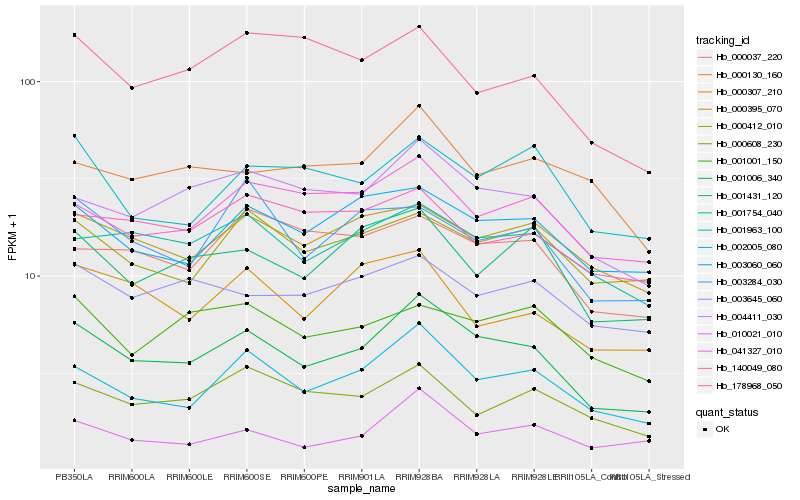
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001006_340 |
0.0 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 7-like [Malus domestica] |
| 2 |
Hb_002005_080 |
0.0805008998 |
- |
- |
PREDICTED: uncharacterized protein LOC105649750 isoform X1 [Jatropha curcas] |
| 3 |
Hb_004411_030 |
0.0897246472 |
- |
- |
hypothetical protein POPTR_0017s00310g [Populus trichocarpa] |
| 4 |
Hb_003060_060 |
0.1068595373 |
- |
- |
PREDICTED: protein transport protein SEC31 homolog B [Jatropha curcas] |
| 5 |
Hb_001431_120 |
0.1091257332 |
- |
- |
PREDICTED: 116 kDa U5 small nuclear ribonucleoprotein component-like [Jatropha curcas] |
| 6 |
Hb_001754_040 |
0.1093112796 |
transcription factor |
TF Family: TRAF |
PREDICTED: uncharacterized protein LOC105643434 [Jatropha curcas] |
| 7 |
Hb_041327_010 |
0.1094647183 |
- |
- |
PREDICTED: la-related protein 1A isoform X1 [Jatropha curcas] |
| 8 |
Hb_000395_070 |
0.1113013518 |
- |
- |
PREDICTED: serine/threonine-protein kinase BLUS1 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000412_010 |
0.1173968594 |
- |
- |
hect ubiquitin-protein ligase, putative [Ricinus communis] |
| 10 |
Hb_000608_230 |
0.1176154867 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 11 |
Hb_001963_100 |
0.1179760746 |
transcription factor |
TF Family: bZIP |
PREDICTED: bZIP transcription factor 17-like [Jatropha curcas] |
| 12 |
Hb_000037_220 |
0.1187473083 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 11 homolog [Jatropha curcas] |
| 13 |
Hb_010021_010 |
0.1189275387 |
- |
- |
unnamed protein product [Coffea canephora] |
| 14 |
Hb_003284_030 |
0.1191815936 |
transcription factor |
TF Family: Orphans |
PREDICTED: paired amphipathic helix protein Sin3-like 2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000307_210 |
0.1209306648 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |
| 16 |
Hb_001001_150 |
0.1218345167 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_178968_050 |
0.1221201143 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |
| 18 |
Hb_003645_060 |
0.1224367824 |
- |
- |
PREDICTED: monodehydroascorbate reductase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_140049_080 |
0.1236098228 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Jatropha curcas] |
| 20 |
Hb_000130_160 |
0.1238899181 |
- |
- |
PREDICTED: protein DJ-1 homolog B-like [Jatropha curcas] |