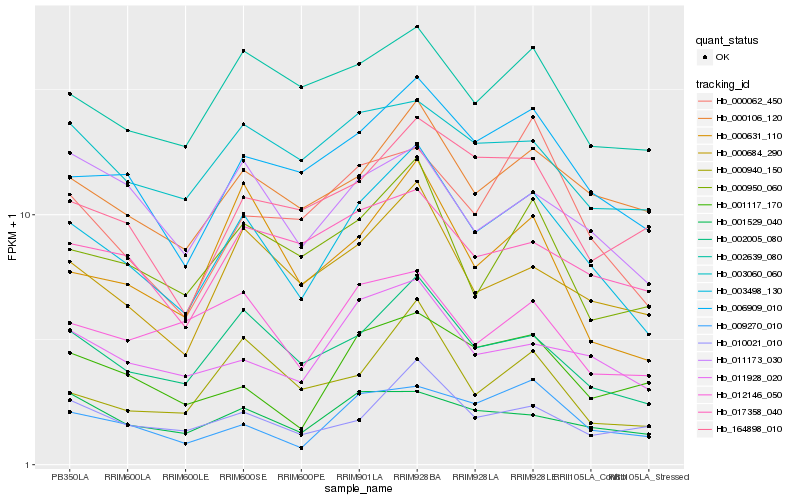
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003498_130 |
0.0 |
- |
- |
PREDICTED: protein ROOT HAIR DEFECTIVE 3 homolog 2-like [Citrus sinensis] |
| 2 |
Hb_002005_080 |
0.0888189642 |
- |
- |
PREDICTED: uncharacterized protein LOC105649750 isoform X1 [Jatropha curcas] |
| 3 |
Hb_006909_010 |
0.0910907972 |
- |
- |
5-oxoprolinase, putative [Ricinus communis] |
| 4 |
Hb_000106_120 |
0.1043662147 |
- |
- |
hypothetical protein glysoja_007050 [Glycine soja] |
| 5 |
Hb_000684_290 |
0.1074929548 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |
| 6 |
Hb_011928_020 |
0.108888074 |
- |
- |
something about silencing protein sas10, putative [Ricinus communis] |
| 7 |
Hb_000950_060 |
0.1149930238 |
- |
- |
PREDICTED: CCAAT/enhancer-binding protein zeta [Jatropha curcas] |
| 8 |
Hb_009270_010 |
0.1172412692 |
- |
- |
PREDICTED: uncharacterized protein LOC103421213 [Malus domestica] |
| 9 |
Hb_010021_010 |
0.1200823676 |
- |
- |
unnamed protein product [Coffea canephora] |
| 10 |
Hb_002639_080 |
0.1203810015 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000631_110 |
0.1209438191 |
- |
- |
PREDICTED: activating signal cointegrator 1 complex subunit 3 [Jatropha curcas] |
| 12 |
Hb_001529_040 |
0.1255606141 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 13 |
Hb_164898_010 |
0.1270459299 |
- |
- |
Cysteine-rich protein, putative [Theobroma cacao] |
| 14 |
Hb_000062_450 |
0.1273541091 |
- |
- |
hypothetical protein MTR_081s0009, partial [Medicago truncatula] |
| 15 |
Hb_017358_040 |
0.1292386544 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 6-like [Jatropha curcas] |
| 16 |
Hb_003060_060 |
0.1294895977 |
- |
- |
PREDICTED: protein transport protein SEC31 homolog B [Jatropha curcas] |
| 17 |
Hb_001117_170 |
0.1298533411 |
- |
- |
- |
| 18 |
Hb_011173_030 |
0.1301537055 |
- |
- |
pumilio, putative [Ricinus communis] |
| 19 |
Hb_000940_150 |
0.1306417486 |
- |
- |
PREDICTED: uncharacterized protein LOC105643779 [Jatropha curcas] |
| 20 |
Hb_012146_050 |
0.1311941139 |
- |
- |
PREDICTED: protein translocase subunit SECA2, chloroplastic [Jatropha curcas] |