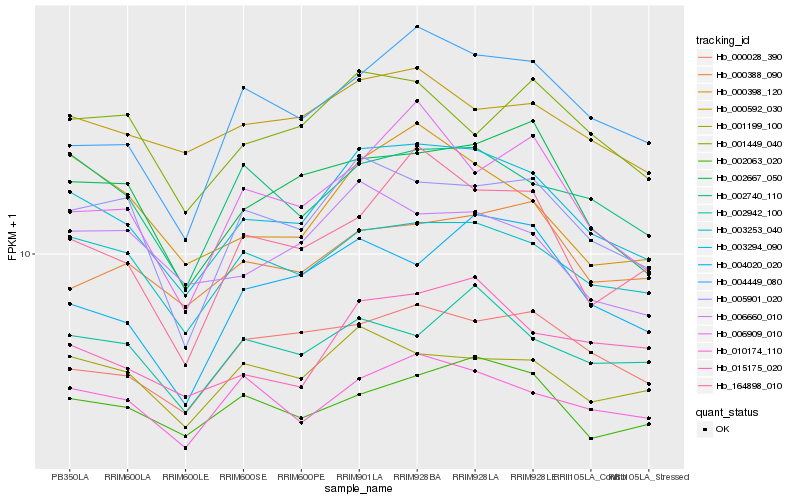
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003294_090 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003253_040 |
0.0697441689 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 3 |
Hb_005901_020 |
0.0760318734 |
- |
- |
PREDICTED: elongation factor Tu GTP-binding domain-containing protein 1 [Jatropha curcas] |
| 4 |
Hb_002740_110 |
0.0837247827 |
- |
- |
PREDICTED: phytochrome A [Jatropha curcas] |
| 5 |
Hb_010174_110 |
0.0837482256 |
- |
- |
hypothetical protein JCGZ_17880 [Jatropha curcas] |
| 6 |
Hb_015175_020 |
0.0845762482 |
- |
- |
hypothetical protein JCGZ_14452 [Jatropha curcas] |
| 7 |
Hb_002063_020 |
0.0866702114 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17140 [Jatropha curcas] |
| 8 |
Hb_002667_050 |
0.0872144663 |
- |
- |
PREDICTED: alpha-mannosidase 2 [Jatropha curcas] |
| 9 |
Hb_001199_100 |
0.0872845674 |
- |
- |
PREDICTED: uncharacterized protein LOC105639813 [Jatropha curcas] |
| 10 |
Hb_004449_080 |
0.0876759239 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH3-like [Jatropha curcas] |
| 11 |
Hb_000398_120 |
0.0877348849 |
- |
- |
PREDICTED: squamous cell carcinoma antigen recognized by T-cells 3-like isoform X4 [Gossypium raimondii] |
| 12 |
Hb_000028_390 |
0.0877376509 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 3-like [Jatropha curcas] |
| 13 |
Hb_000592_030 |
0.0877560454 |
- |
- |
PREDICTED: uncharacterized protein LOC100853969 isoform X1 [Vitis vinifera] |
| 14 |
Hb_006660_010 |
0.0890901513 |
- |
- |
PREDICTED: exocyst complex component EXO84C isoform X1 [Jatropha curcas] |
| 15 |
Hb_164898_010 |
0.0918366299 |
- |
- |
Cysteine-rich protein, putative [Theobroma cacao] |
| 16 |
Hb_002942_100 |
0.0919317478 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g61990, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000388_090 |
0.0935258214 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g77360, mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_006909_010 |
0.0939878613 |
- |
- |
5-oxoprolinase, putative [Ricinus communis] |
| 19 |
Hb_001449_040 |
0.0947591741 |
- |
- |
PREDICTED: SWR1 complex subunit 2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_004020_020 |
0.0954995888 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g65820 [Jatropha curcas] |