| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
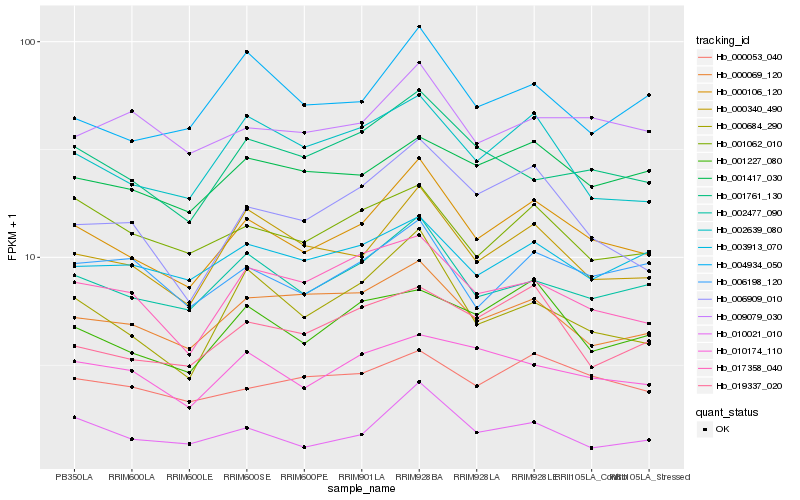
Hb_000106_120 |
0.0 |
- |
- |
hypothetical protein glysoja_007050 [Glycine soja] |
| 2 |
Hb_002477_090 |
0.0739421698 |
- |
- |
PREDICTED: uncharacterized protein LOC105631409 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000340_490 |
0.0747098877 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |
| 4 |
Hb_006909_010 |
0.0863002399 |
- |
- |
5-oxoprolinase, putative [Ricinus communis] |
| 5 |
Hb_002639_080 |
0.0879039871 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000684_290 |
0.0891830833 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |
| 7 |
Hb_000069_120 |
0.090011718 |
- |
- |
PREDICTED: uncharacterized protein LOC105124742 isoform X1 [Populus euphratica] |
| 8 |
Hb_000053_040 |
0.0903475434 |
- |
- |
hypothetical protein CISIN_1g0010392mg, partial [Citrus sinensis] |
| 9 |
Hb_017358_040 |
0.0912250291 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 6-like [Jatropha curcas] |
| 10 |
Hb_001227_080 |
0.0931210135 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_001761_130 |
0.0933695842 |
- |
- |
PREDICTED: uncharacterized protein LOC105646118 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001417_030 |
0.0947576237 |
- |
- |
PREDICTED: inactive poly [ADP-ribose] polymerase RCD1 [Jatropha curcas] |
| 13 |
Hb_006198_120 |
0.0952499001 |
- |
- |
PREDICTED: guanine nucleotide-binding protein-like NSN1 [Jatropha curcas] |
| 14 |
Hb_009079_030 |
0.0955582252 |
- |
- |
PREDICTED: general transcription factor IIF subunit 2 [Jatropha curcas] |
| 15 |
Hb_004934_050 |
0.0957439016 |
- |
- |
Eukaryotic release factor 1-3 isoform 1 [Theobroma cacao] |
| 16 |
Hb_001062_010 |
0.0962338579 |
- |
- |
PREDICTED: CWF19-like protein 2 [Jatropha curcas] |
| 17 |
Hb_010021_010 |
0.0962982052 |
- |
- |
unnamed protein product [Coffea canephora] |
| 18 |
Hb_010174_110 |
0.097139607 |
- |
- |
hypothetical protein JCGZ_17880 [Jatropha curcas] |
| 19 |
Hb_003913_070 |
0.0984923537 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 30 [Populus euphratica] |
| 20 |
Hb_019337_020 |
0.0989704752 |
- |
- |
K+ uptake permease 11 isoform 1 [Theobroma cacao] |