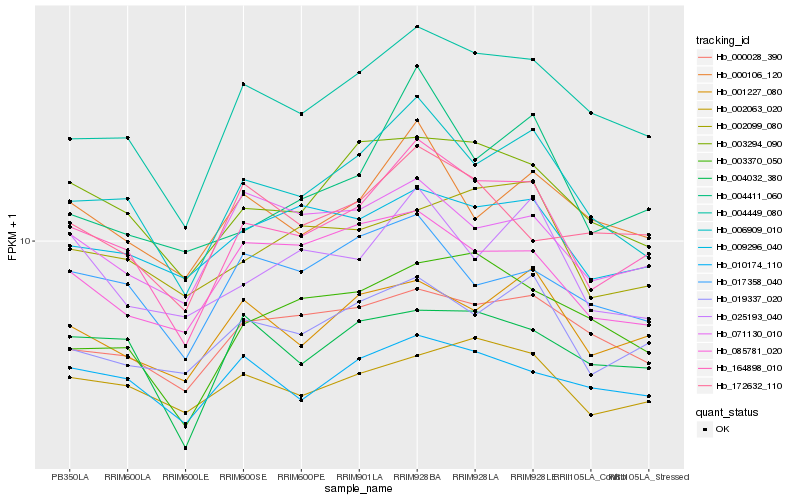
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_164898_010 |
0.0 |
- |
- |
Cysteine-rich protein, putative [Theobroma cacao] |
| 2 |
Hb_006909_010 |
0.0704164119 |
- |
- |
5-oxoprolinase, putative [Ricinus communis] |
| 3 |
Hb_004449_080 |
0.07504217 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH3-like [Jatropha curcas] |
| 4 |
Hb_003294_090 |
0.0918366299 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_019337_020 |
0.095287526 |
- |
- |
K+ uptake permease 11 isoform 1 [Theobroma cacao] |
| 6 |
Hb_001227_080 |
0.1027929327 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_000106_120 |
0.1044144895 |
- |
- |
hypothetical protein glysoja_007050 [Glycine soja] |
| 8 |
Hb_000028_390 |
0.1083448917 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 3-like [Jatropha curcas] |
| 9 |
Hb_003370_050 |
0.108668071 |
- |
- |
PREDICTED: uncharacterized protein LOC103697550 [Phoenix dactylifera] |
| 10 |
Hb_004411_060 |
0.108838234 |
- |
- |
acetolactate synthase, putative [Ricinus communis] |
| 11 |
Hb_002063_020 |
0.1090834556 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17140 [Jatropha curcas] |
| 12 |
Hb_004032_380 |
0.1090985838 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5-like [Jatropha curcas] |
| 13 |
Hb_009296_040 |
0.1096402447 |
- |
- |
hypothetical protein L484_007435 [Morus notabilis] |
| 14 |
Hb_010174_110 |
0.1103921858 |
- |
- |
hypothetical protein JCGZ_17880 [Jatropha curcas] |
| 15 |
Hb_085781_020 |
0.1103993721 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1 [Jatropha curcas] |
| 16 |
Hb_071130_010 |
0.1119127427 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_172632_110 |
0.1122461944 |
- |
- |
PREDICTED: pumilio homolog 5 [Jatropha curcas] |
| 18 |
Hb_025193_040 |
0.1129329268 |
- |
- |
PREDICTED: uncharacterized protein LOC105647286 [Jatropha curcas] |
| 19 |
Hb_017358_040 |
0.1139919386 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 6-like [Jatropha curcas] |
| 20 |
Hb_002099_080 |
0.1143071379 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 37-like [Jatropha curcas] |