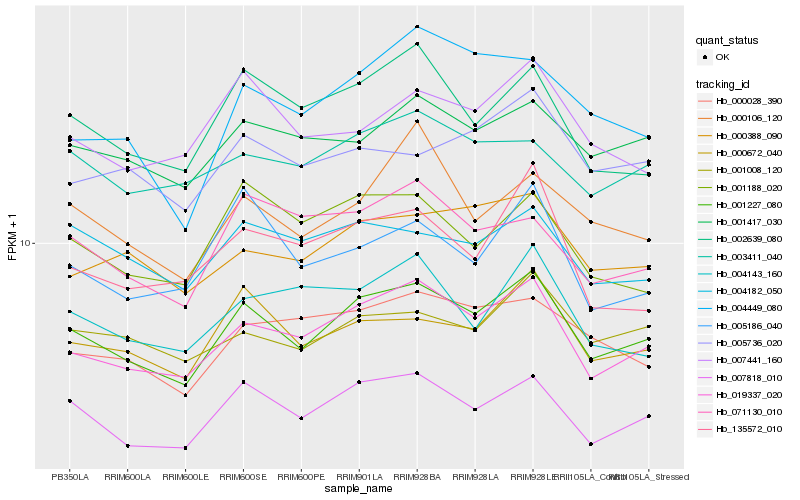
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001227_080 |
0.0 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_019337_020 |
0.0478641654 |
- |
- |
K+ uptake permease 11 isoform 1 [Theobroma cacao] |
| 3 |
Hb_007818_010 |
0.0536901622 |
- |
- |
PREDICTED: transcriptional corepressor SEUSS-like isoform X2 [Gossypium raimondii] |
| 4 |
Hb_000672_040 |
0.0708802303 |
- |
- |
PREDICTED: ENHANCER OF AG-4 protein 2 [Jatropha curcas] |
| 5 |
Hb_002639_080 |
0.0806389026 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001417_030 |
0.0820646913 |
- |
- |
PREDICTED: inactive poly [ADP-ribose] polymerase RCD1 [Jatropha curcas] |
| 7 |
Hb_000388_090 |
0.0820955963 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g77360, mitochondrial-like [Jatropha curcas] |
| 8 |
Hb_001008_120 |
0.08338663 |
desease resistance |
Gene Name: PEX-1N |
peroxisome biogenesis factor, putative [Ricinus communis] |
| 9 |
Hb_000028_390 |
0.0863343291 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 3-like [Jatropha curcas] |
| 10 |
Hb_007441_160 |
0.0880145962 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_004182_050 |
0.0888950231 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL3 isoform X2 [Jatropha curcas] |
| 12 |
Hb_003411_040 |
0.0889891435 |
- |
- |
unnamed protein product [Coffea canephora] |
| 13 |
Hb_001188_020 |
0.0894318602 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_071130_010 |
0.089794178 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_005736_020 |
0.0915024405 |
- |
- |
hypothetical protein POPTR_0005s20940g [Populus trichocarpa] |
| 16 |
Hb_135572_010 |
0.0919929411 |
- |
- |
GTP-binding protein alpha subunit, gna, putative [Ricinus communis] |
| 17 |
Hb_004449_080 |
0.0927612735 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH3-like [Jatropha curcas] |
| 18 |
Hb_000106_120 |
0.0931210135 |
- |
- |
hypothetical protein glysoja_007050 [Glycine soja] |
| 19 |
Hb_005186_040 |
0.0931829692 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 13 isoform X2 [Jatropha curcas] |
| 20 |
Hb_004143_160 |
0.0932554602 |
- |
- |
SAB, putative [Ricinus communis] |