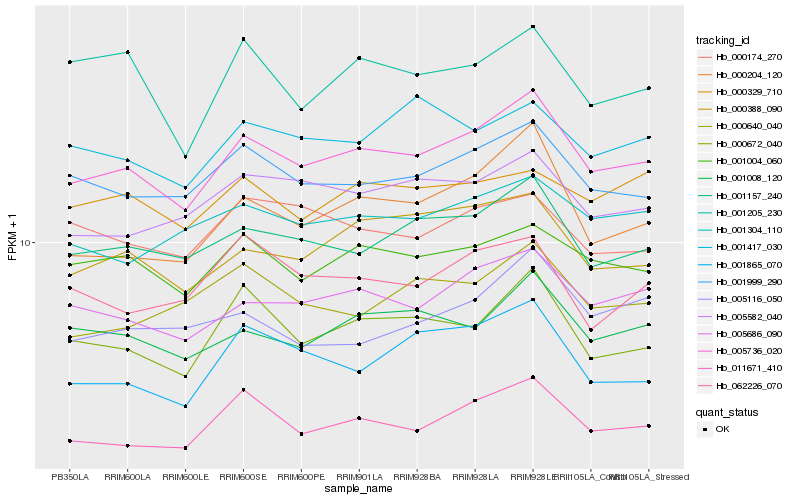
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005736_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0005s20940g [Populus trichocarpa] |
| 2 |
Hb_005686_090 |
0.0517674289 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001157_240 |
0.0554805203 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 37-like [Jatropha curcas] |
| 4 |
Hb_001999_290 |
0.0574977414 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 7, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_001004_060 |
0.0614495903 |
- |
- |
PREDICTED: uncharacterized protein LOC105630220 isoform X4 [Jatropha curcas] |
| 6 |
Hb_001008_120 |
0.063766945 |
desease resistance |
Gene Name: PEX-1N |
peroxisome biogenesis factor, putative [Ricinus communis] |
| 7 |
Hb_000204_120 |
0.0644044992 |
- |
- |
PREDICTED: uncharacterized protein LOC105639239 [Jatropha curcas] |
| 8 |
Hb_000388_090 |
0.065423552 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g77360, mitochondrial-like [Jatropha curcas] |
| 9 |
Hb_000672_040 |
0.0658471852 |
- |
- |
PREDICTED: ENHANCER OF AG-4 protein 2 [Jatropha curcas] |
| 10 |
Hb_011671_410 |
0.0666783733 |
transcription factor |
TF Family: SET |
histone-lysine n-methyltransferase, suvh, putative [Ricinus communis] |
| 11 |
Hb_005582_040 |
0.0687263257 |
- |
- |
PREDICTED: regulator of nonsense transcripts 1 homolog [Jatropha curcas] |
| 12 |
Hb_001304_110 |
0.0702803154 |
transcription factor |
TF Family: WRKY |
WRKY protein [Hevea brasiliensis] |
| 13 |
Hb_000640_040 |
0.0773223536 |
- |
- |
PREDICTED: bromodomain and WD repeat-containing protein 1 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000329_710 |
0.0786107961 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase BRE1-like 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001417_030 |
0.079144458 |
- |
- |
PREDICTED: inactive poly [ADP-ribose] polymerase RCD1 [Jatropha curcas] |
| 16 |
Hb_001865_070 |
0.0792666824 |
- |
- |
PREDICTED: uncharacterized protein LOC105647937 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000174_270 |
0.0803151403 |
- |
- |
PREDICTED: type II inositol 1,4,5-trisphosphate 5-phosphatase FRA3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_062226_070 |
0.0813365851 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 4 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001205_230 |
0.0818447852 |
- |
- |
PREDICTED: acidic leucine-rich nuclear phosphoprotein 32-related protein [Jatropha curcas] |
| 20 |
Hb_005116_050 |
0.0820913193 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02820, mitochondrial isoform X1 [Jatropha curcas] |