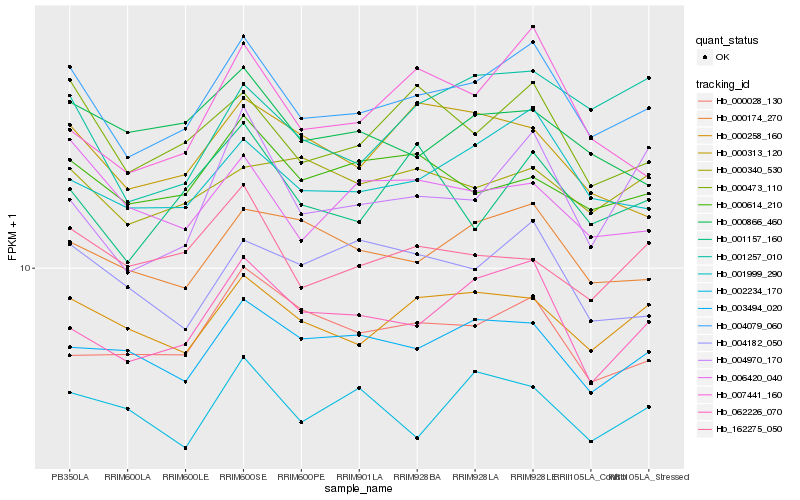
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004079_060 |
0.0 |
- |
- |
hypothetical protein JCGZ_07228 [Jatropha curcas] |
| 2 |
Hb_001999_290 |
0.0547215426 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 7, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_062226_070 |
0.0618631732 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 4 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000473_110 |
0.0620879583 |
- |
- |
PREDICTED: PP2A regulatory subunit TAP46 [Jatropha curcas] |
| 5 |
Hb_000258_160 |
0.0639556279 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000174_270 |
0.0681627324 |
- |
- |
PREDICTED: type II inositol 1,4,5-trisphosphate 5-phosphatase FRA3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003494_020 |
0.0724385392 |
- |
- |
ATP-dependent RNA and DNA helicase, putative [Ricinus communis] |
| 8 |
Hb_000614_210 |
0.076562846 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_006420_040 |
0.0779043458 |
transcription factor |
TF Family: Orphans |
PREDICTED: paired amphipathic helix protein Sin3-like 4 isoform X2 [Jatropha curcas] |
| 10 |
Hb_004970_170 |
0.0779683576 |
transcription factor |
TF Family: bZIP |
PREDICTED: ABSCISIC ACID-INSENSITIVE 5-like protein 5 [Jatropha curcas] |
| 11 |
Hb_000028_130 |
0.0780166515 |
- |
- |
PREDICTED: uncharacterized protein LOC105645303 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001157_160 |
0.0783029962 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase BRE1-like 2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_004182_050 |
0.0795657897 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL3 isoform X2 [Jatropha curcas] |
| 14 |
Hb_162275_050 |
0.0800378889 |
- |
- |
PREDICTED: mRNA-capping enzyme [Jatropha curcas] |
| 15 |
Hb_007441_160 |
0.0803153698 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_000866_460 |
0.080742416 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_000340_530 |
0.0811349673 |
- |
- |
hypothetical protein VITISV_016664 [Vitis vinifera] |
| 18 |
Hb_000313_120 |
0.0812294127 |
- |
- |
PREDICTED: ALG-2 interacting protein X [Jatropha curcas] |
| 19 |
Hb_002234_170 |
0.0820505097 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_001257_010 |
0.083104293 |
- |
- |
PREDICTED: apoptosis-inducing factor 2-like isoform X1 [Jatropha curcas] |