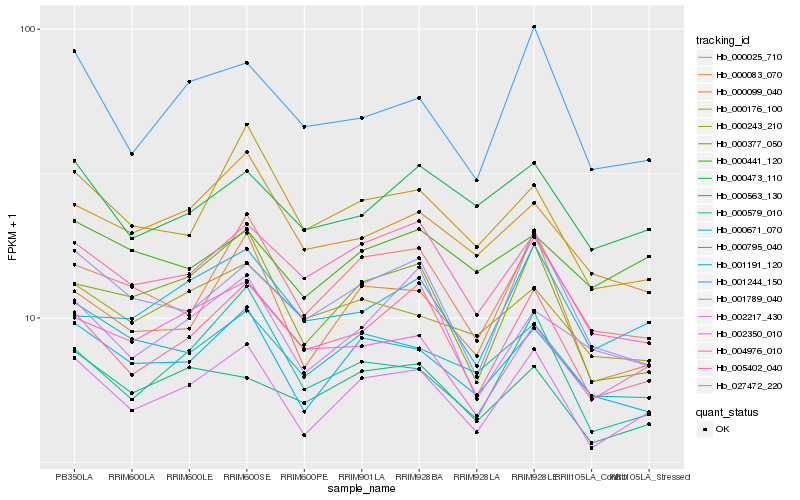
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002350_010 |
0.0 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit rpc1 [Jatropha curcas] |
| 2 |
Hb_001191_120 |
0.0527834523 |
- |
- |
PREDICTED: cleavage stimulating factor 64 isoform X1 [Jatropha curcas] |
| 3 |
Hb_027472_220 |
0.0650129458 |
- |
- |
PREDICTED: nucleolar complex protein 3 homolog isoform X1 [Jatropha curcas] |
| 4 |
Hb_000671_070 |
0.0699514047 |
- |
- |
PREDICTED: uncharacterized protein LOC105630621 [Jatropha curcas] |
| 5 |
Hb_000025_710 |
0.0700866628 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 6 |
Hb_000473_110 |
0.0722571187 |
- |
- |
PREDICTED: PP2A regulatory subunit TAP46 [Jatropha curcas] |
| 7 |
Hb_000563_130 |
0.0732235597 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 8 |
Hb_005402_040 |
0.073309569 |
- |
- |
PREDICTED: replication factor C subunit 1 [Jatropha curcas] |
| 9 |
Hb_000243_210 |
0.074427865 |
- |
- |
protein with unknown function [Ricinus communis] |
| 10 |
Hb_000083_070 |
0.0756197034 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 1 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000579_010 |
0.0756976217 |
transcription factor |
TF Family: SET |
PREDICTED: probable histone-lysine N-methyltransferase ATXR3 [Jatropha curcas] |
| 12 |
Hb_000377_050 |
0.0775141596 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 13 |
Hb_000795_040 |
0.0776708446 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 14 |
Hb_000441_120 |
0.077841528 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT111, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_001244_150 |
0.0782359676 |
- |
- |
transformer serine/arginine-rich ribonucleoprotein [Populus trichocarpa] |
| 16 |
Hb_000099_040 |
0.0784003056 |
- |
- |
unnamed protein product [Coffea canephora] |
| 17 |
Hb_001789_040 |
0.078676628 |
- |
- |
PREDICTED: uncharacterized protein LOC105648457 isoform X2 [Jatropha curcas] |
| 18 |
Hb_002217_430 |
0.0806599527 |
- |
- |
PREDICTED: flowering time control protein FCA isoform X4 [Jatropha curcas] |
| 19 |
Hb_004976_010 |
0.0807735828 |
- |
- |
hypothetical protein RCOM_1429110 [Ricinus communis] |
| 20 |
Hb_000176_100 |
0.0811836658 |
- |
- |
PREDICTED: probable splicing factor 3A subunit 1 [Solanum lycopersicum] |