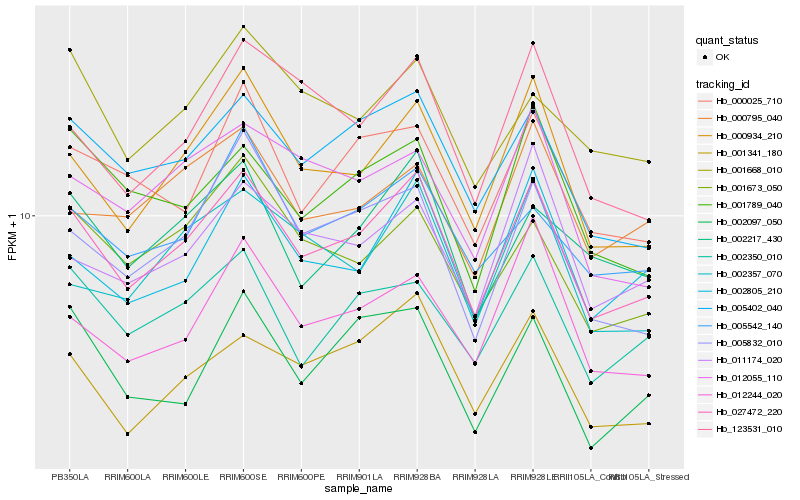
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027472_220 |
0.0 |
- |
- |
PREDICTED: nucleolar complex protein 3 homolog isoform X1 [Jatropha curcas] |
| 2 |
Hb_000934_210 |
0.0504158595 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 3 |
Hb_005402_040 |
0.0511046361 |
- |
- |
PREDICTED: replication factor C subunit 1 [Jatropha curcas] |
| 4 |
Hb_001341_180 |
0.0589304675 |
- |
- |
PREDICTED: transmembrane protein 209 [Jatropha curcas] |
| 5 |
Hb_002805_210 |
0.0633398177 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 6 |
Hb_002350_010 |
0.0650129458 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit rpc1 [Jatropha curcas] |
| 7 |
Hb_005832_010 |
0.0668326505 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 8 |
Hb_005542_140 |
0.0675182713 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 9 |
Hb_002217_430 |
0.0676328102 |
- |
- |
PREDICTED: flowering time control protein FCA isoform X4 [Jatropha curcas] |
| 10 |
Hb_001673_050 |
0.0685848546 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000025_710 |
0.0686425775 |
transcription factor |
TF Family: SNF2 |
Chromodomain-helicase-DNA-binding 2 [Gossypium arboreum] |
| 12 |
Hb_123531_010 |
0.0707381478 |
- |
- |
PREDICTED: nuclear pore complex protein NUP98A isoform X1 [Jatropha curcas] |
| 13 |
Hb_012244_020 |
0.0714202601 |
- |
- |
calpain, putative [Ricinus communis] |
| 14 |
Hb_001789_040 |
0.0725249803 |
- |
- |
PREDICTED: uncharacterized protein LOC105648457 isoform X2 [Jatropha curcas] |
| 15 |
Hb_011174_020 |
0.072886933 |
- |
- |
PREDICTED: uncharacterized protein LOC105632610 [Jatropha curcas] |
| 16 |
Hb_012055_110 |
0.0729335207 |
- |
- |
tetratricopeptide repeat protein, tpr, putative [Ricinus communis] |
| 17 |
Hb_002097_050 |
0.0732970609 |
- |
- |
PREDICTED: protein fluG isoform X1 [Jatropha curcas] |
| 18 |
Hb_001668_010 |
0.0733572206 |
- |
- |
PREDICTED: RNA-binding protein with multiple splicing [Jatropha curcas] |
| 19 |
Hb_002357_070 |
0.0752771756 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 20 |
Hb_000795_040 |
0.0761053553 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |