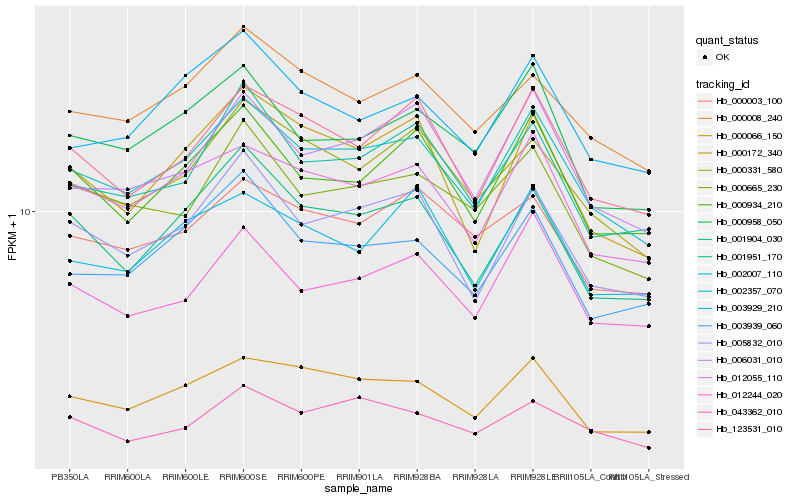
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002007_110 |
0.0 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X2 [Jatropha curcas] |
| 2 |
Hb_012055_110 |
0.0478311855 |
- |
- |
tetratricopeptide repeat protein, tpr, putative [Ricinus communis] |
| 3 |
Hb_123531_010 |
0.0525254839 |
- |
- |
PREDICTED: nuclear pore complex protein NUP98A isoform X1 [Jatropha curcas] |
| 4 |
Hb_005832_010 |
0.0531701508 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000331_580 |
0.0547387313 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 6 |
Hb_000066_150 |
0.0551600263 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 7 |
Hb_006031_010 |
0.0555493516 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15b isoform X3 [Jatropha curcas] |
| 8 |
Hb_000934_210 |
0.0556618555 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 9 |
Hb_000008_240 |
0.0577756224 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001951_170 |
0.0595736604 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 11 |
Hb_000958_050 |
0.0604109542 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |
| 12 |
Hb_000665_230 |
0.0607870568 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 13 |
Hb_012244_020 |
0.0613828614 |
- |
- |
calpain, putative [Ricinus communis] |
| 14 |
Hb_001904_030 |
0.0619418979 |
- |
- |
PREDICTED: importin-5 [Jatropha curcas] |
| 15 |
Hb_003939_060 |
0.0634829104 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000003_100 |
0.0644082453 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |
| 17 |
Hb_003929_210 |
0.065162978 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 18 |
Hb_000172_340 |
0.066943092 |
- |
- |
PREDICTED: uncharacterized protein LOC101261327 [Solanum lycopersicum] |
| 19 |
Hb_002357_070 |
0.0686589945 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 20 |
Hb_043362_010 |
0.0687755259 |
- |
- |
hypothetical protein JCGZ_23765 [Jatropha curcas] |