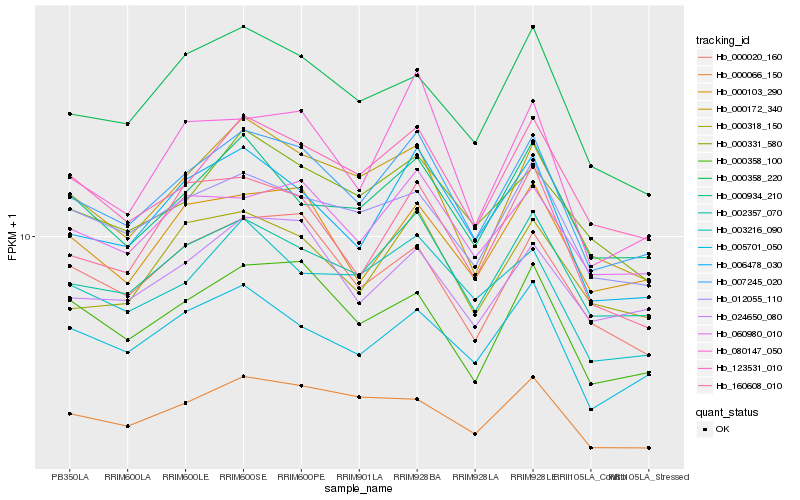
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007245_020 |
0.0 |
- |
- |
PREDICTED: pre-mRNA-processing factor 17-like isoform X1 [Populus euphratica] |
| 2 |
Hb_002357_070 |
0.046028484 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 3 |
Hb_123531_010 |
0.0529390145 |
- |
- |
PREDICTED: nuclear pore complex protein NUP98A isoform X1 [Jatropha curcas] |
| 4 |
Hb_006478_030 |
0.0574719865 |
- |
- |
hypothetical protein POPTR_0005s23510g [Populus trichocarpa] |
| 5 |
Hb_000172_340 |
0.0628083587 |
- |
- |
PREDICTED: uncharacterized protein LOC101261327 [Solanum lycopersicum] |
| 6 |
Hb_060980_010 |
0.064585299 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 7 |
Hb_000103_290 |
0.068166321 |
- |
- |
PREDICTED: dymeclin isoform X1 [Jatropha curcas] |
| 8 |
Hb_000358_100 |
0.0696258834 |
- |
- |
PREDICTED: uncharacterized protein LOC105633371 [Jatropha curcas] |
| 9 |
Hb_012055_110 |
0.0699232141 |
- |
- |
tetratricopeptide repeat protein, tpr, putative [Ricinus communis] |
| 10 |
Hb_000934_210 |
0.0712085383 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 11 |
Hb_005701_050 |
0.0712181212 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 12 |
Hb_000331_580 |
0.0712802824 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 13 |
Hb_024650_080 |
0.0720961293 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g09030 [Jatropha curcas] |
| 14 |
Hb_003216_090 |
0.0722964368 |
- |
- |
PREDICTED: nuclear pore complex protein NUP133 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000066_150 |
0.0724000616 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 16 |
Hb_000318_150 |
0.0733103043 |
- |
- |
RNA-binding protein Nova-1, putative [Ricinus communis] |
| 17 |
Hb_000020_160 |
0.0734254093 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1-like [Gossypium raimondii] |
| 18 |
Hb_160608_010 |
0.0738660981 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 19 |
Hb_080147_050 |
0.0741584811 |
- |
- |
PREDICTED: probable plastidic glucose transporter 2 [Jatropha curcas] |
| 20 |
Hb_000358_220 |
0.074321466 |
- |
- |
PREDICTED: SNW/SKI-interacting protein-like [Jatropha curcas] |