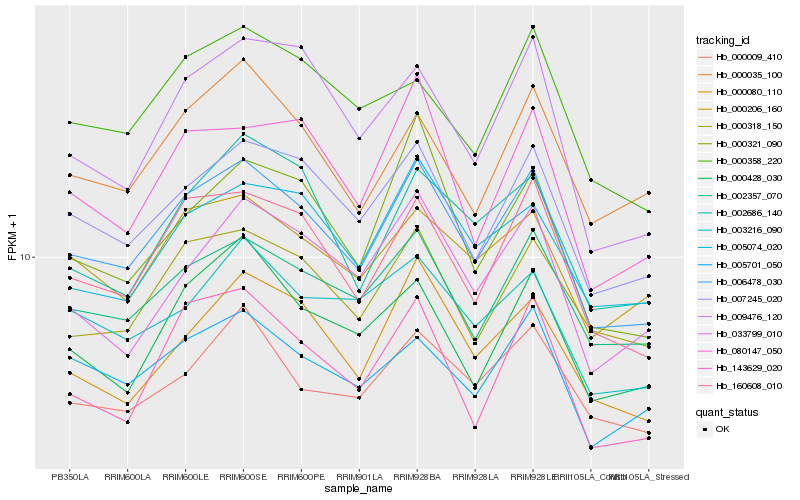
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006478_030 |
0.0 |
- |
- |
hypothetical protein POPTR_0005s23510g [Populus trichocarpa] |
| 2 |
Hb_160608_010 |
0.0533351721 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 3 |
Hb_007245_020 |
0.0574719865 |
- |
- |
PREDICTED: pre-mRNA-processing factor 17-like isoform X1 [Populus euphratica] |
| 4 |
Hb_002357_070 |
0.0643087583 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 5 |
Hb_000318_150 |
0.0706285676 |
- |
- |
RNA-binding protein Nova-1, putative [Ricinus communis] |
| 6 |
Hb_000080_110 |
0.073687446 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000206_160 |
0.0758802624 |
- |
- |
PREDICTED: TBCC domain-containing protein 1 [Jatropha curcas] |
| 8 |
Hb_005701_050 |
0.0761891092 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 9 |
Hb_005074_020 |
0.07751206 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 10 |
Hb_080147_050 |
0.0778366845 |
- |
- |
PREDICTED: probable plastidic glucose transporter 2 [Jatropha curcas] |
| 11 |
Hb_033799_010 |
0.0803920467 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |
| 12 |
Hb_009476_120 |
0.0804981457 |
- |
- |
Clathrin heavy chain 1 [Glycine soja] |
| 13 |
Hb_143629_020 |
0.0805168031 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 14 |
Hb_000428_030 |
0.0805271952 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 15 |
Hb_000321_090 |
0.0807434289 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 16 |
Hb_000009_410 |
0.0807855861 |
transcription factor |
TF Family: SET |
hypothetical protein POPTR_0007s12130g [Populus trichocarpa] |
| 17 |
Hb_003216_090 |
0.0816534668 |
- |
- |
PREDICTED: nuclear pore complex protein NUP133 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000035_100 |
0.0818280767 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 19 |
Hb_002686_140 |
0.081828341 |
- |
- |
PREDICTED: general negative regulator of transcription subunit 3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000358_220 |
0.0838045224 |
- |
- |
PREDICTED: SNW/SKI-interacting protein-like [Jatropha curcas] |