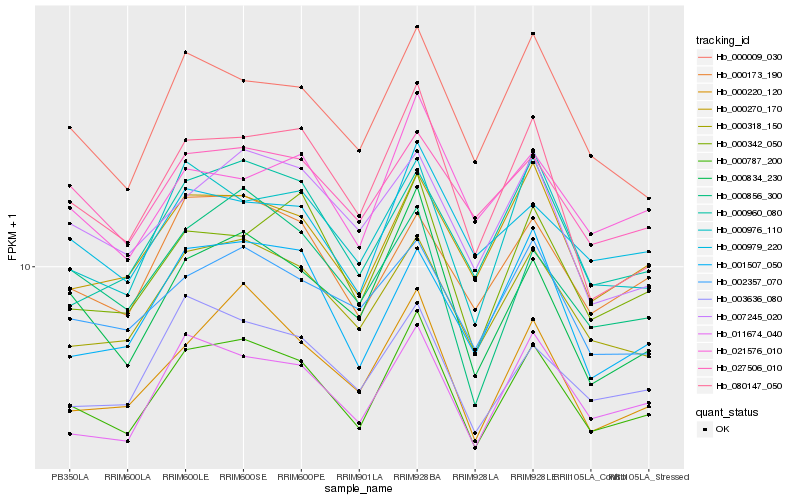
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000787_200 |
0.0 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_000318_150 |
0.0626576494 |
- |
- |
RNA-binding protein Nova-1, putative [Ricinus communis] |
| 3 |
Hb_011674_040 |
0.0644199043 |
- |
- |
PREDICTED: uncharacterized protein LOC105648163 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000834_230 |
0.0693077088 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |
| 5 |
Hb_080147_050 |
0.0730975036 |
- |
- |
PREDICTED: probable plastidic glucose transporter 2 [Jatropha curcas] |
| 6 |
Hb_000342_050 |
0.0740615377 |
- |
- |
hypothetical protein POPTR_0006s24710g [Populus trichocarpa] |
| 7 |
Hb_000270_170 |
0.0742746432 |
- |
- |
PREDICTED: formin-like protein 5 [Jatropha curcas] |
| 8 |
Hb_000220_120 |
0.0746799982 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000173_190 |
0.0757940433 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003636_080 |
0.0769352825 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80B1 [Jatropha curcas] |
| 11 |
Hb_000976_110 |
0.0771969639 |
- |
- |
PREDICTED: KRR1 small subunit processome component homolog [Jatropha curcas] |
| 12 |
Hb_000009_030 |
0.0773657453 |
- |
- |
PREDICTED: splicing factor U2af large subunit B isoform X1 [Jatropha curcas] |
| 13 |
Hb_002357_070 |
0.0797275821 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 14 |
Hb_000979_220 |
0.0805259476 |
- |
- |
PREDICTED: uncharacterized protein LOC105634085 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000960_080 |
0.0825168518 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_001507_050 |
0.0847754379 |
- |
- |
PREDICTED: cysteine desulfurase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_027506_010 |
0.0856199666 |
- |
- |
PREDICTED: cullin-4 [Jatropha curcas] |
| 18 |
Hb_007245_020 |
0.0859273186 |
- |
- |
PREDICTED: pre-mRNA-processing factor 17-like isoform X1 [Populus euphratica] |
| 19 |
Hb_021576_010 |
0.0861345089 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 20 |
Hb_000856_300 |
0.0881014719 |
- |
- |
PREDICTED: uncharacterized protein LOC105640498 [Jatropha curcas] |