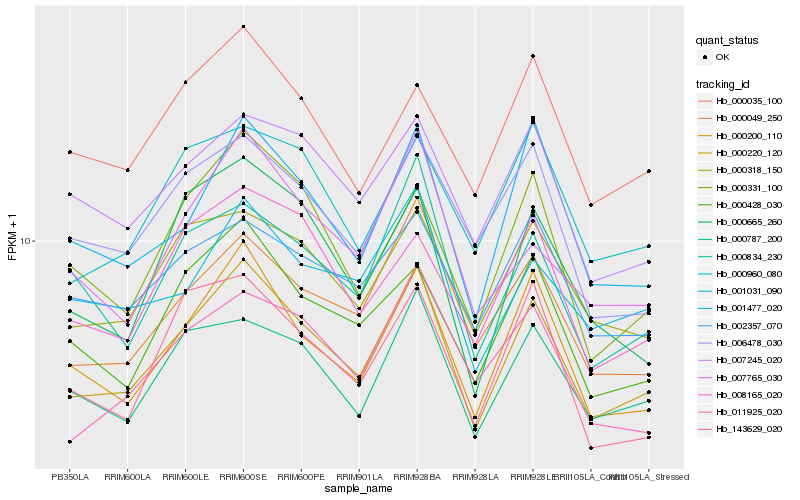
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000220_120 |
0.0 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000200_110 |
0.0621377786 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000049_250 |
0.0732199569 |
- |
- |
PREDICTED: uncharacterized protein LOC105644455 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000787_200 |
0.0746799982 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_001477_020 |
0.0754514044 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105646886 isoform X3 [Jatropha curcas] |
| 6 |
Hb_000428_030 |
0.0783649228 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 7 |
Hb_000318_150 |
0.0796812102 |
- |
- |
RNA-binding protein Nova-1, putative [Ricinus communis] |
| 8 |
Hb_000331_100 |
0.0848114959 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 9 |
Hb_007245_020 |
0.0873596775 |
- |
- |
PREDICTED: pre-mRNA-processing factor 17-like isoform X1 [Populus euphratica] |
| 10 |
Hb_006478_030 |
0.0878092324 |
- |
- |
hypothetical protein POPTR_0005s23510g [Populus trichocarpa] |
| 11 |
Hb_000834_230 |
0.0880870257 |
- |
- |
PREDICTED: ras-related protein RHN1-like [Jatropha curcas] |
| 12 |
Hb_000960_080 |
0.0883335639 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_008165_020 |
0.088871364 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 14 |
Hb_007765_030 |
0.0890435285 |
- |
- |
PREDICTED: neutral ceramidase [Jatropha curcas] |
| 15 |
Hb_001031_090 |
0.0891713378 |
- |
- |
PREDICTED: translational activator GCN1 [Jatropha curcas] |
| 16 |
Hb_011925_020 |
0.0897641341 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 17 |
Hb_143629_020 |
0.0899163466 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 18 |
Hb_002357_070 |
0.0901207011 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 19 |
Hb_000665_260 |
0.0906072572 |
- |
- |
PREDICTED: uncharacterized protein LOC105637620 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000035_100 |
0.090971805 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |