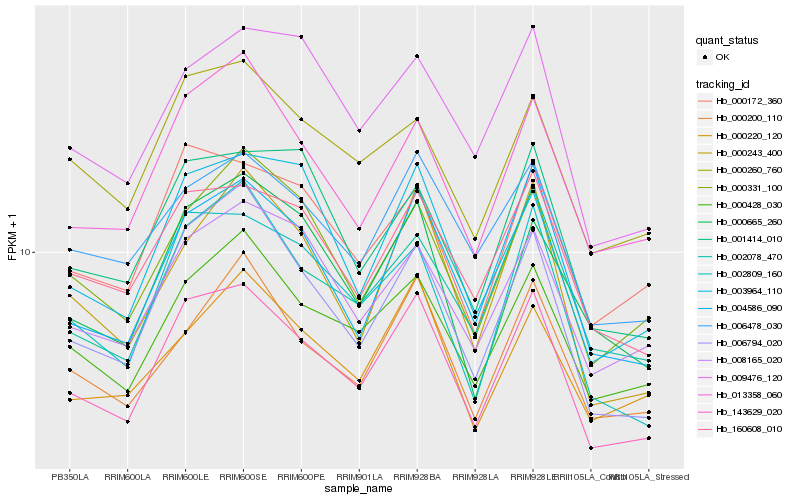
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_143629_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 2 |
Hb_004586_090 |
0.0655930947 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 16 isoform X1 [Jatropha curcas] |
| 3 |
Hb_006794_020 |
0.0685650822 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X6 [Jatropha curcas] |
| 4 |
Hb_000428_030 |
0.0706800669 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 5 |
Hb_160608_010 |
0.0715295511 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |
| 6 |
Hb_002809_160 |
0.0720938868 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 7 |
Hb_002078_470 |
0.07368171 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 4 isoform X1 [Jatropha curcas] |
| 8 |
Hb_013358_060 |
0.0762737866 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |
| 9 |
Hb_000172_360 |
0.0792282645 |
- |
- |
PREDICTED: apoptotic chromatin condensation inducer in the nucleus [Jatropha curcas] |
| 10 |
Hb_000331_100 |
0.0802393124 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 11 |
Hb_006478_030 |
0.0805168031 |
- |
- |
hypothetical protein POPTR_0005s23510g [Populus trichocarpa] |
| 12 |
Hb_000200_110 |
0.0807126636 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003964_110 |
0.081539662 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g26460, mitochondrial [Jatropha curcas] |
| 14 |
Hb_000243_400 |
0.082004231 |
- |
- |
PREDICTED: nucleolin-like [Jatropha curcas] |
| 15 |
Hb_000665_260 |
0.0832180333 |
- |
- |
PREDICTED: uncharacterized protein LOC105637620 isoform X1 [Jatropha curcas] |
| 16 |
Hb_008165_020 |
0.0837000669 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000260_760 |
0.0860300139 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 18 |
Hb_001414_010 |
0.0877587172 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 19 |
Hb_009476_120 |
0.0889580422 |
- |
- |
Clathrin heavy chain 1 [Glycine soja] |
| 20 |
Hb_000220_120 |
0.0899163466 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 isoform X2 [Jatropha curcas] |