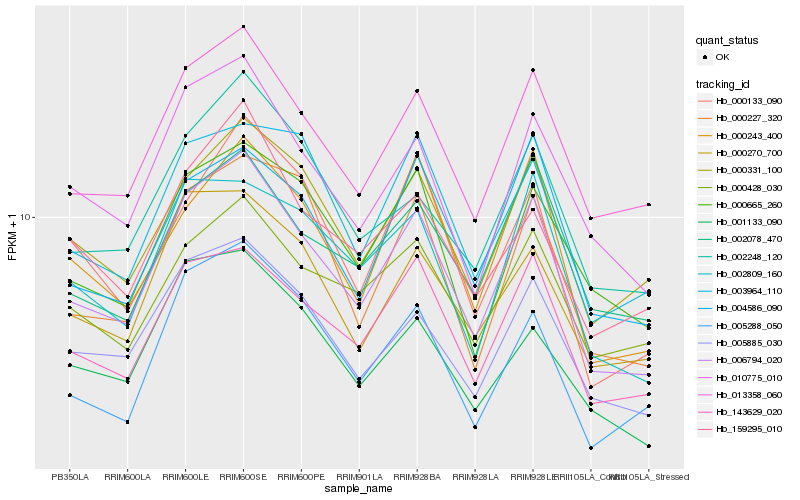
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006794_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X6 [Jatropha curcas] |
| 2 |
Hb_013358_060 |
0.0675421503 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |
| 3 |
Hb_143629_020 |
0.0685650822 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |
| 4 |
Hb_005885_030 |
0.06988831 |
desease resistance |
Gene Name: TIR |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 5 |
Hb_010775_010 |
0.0748848158 |
- |
- |
PREDICTED: uncharacterized protein LOC105642163 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000331_100 |
0.0819261387 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000270_700 |
0.0857363981 |
- |
- |
PREDICTED: protease Do-like 9 [Populus euphratica] |
| 8 |
Hb_000243_400 |
0.0878014435 |
- |
- |
PREDICTED: nucleolin-like [Jatropha curcas] |
| 9 |
Hb_002809_160 |
0.0882587303 |
- |
- |
PREDICTED: protein FAM63A [Jatropha curcas] |
| 10 |
Hb_002248_120 |
0.0889733493 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 11 |
Hb_000227_320 |
0.0896247987 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_002078_470 |
0.0896384263 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 4 isoform X1 [Jatropha curcas] |
| 13 |
Hb_005288_050 |
0.0900811353 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR4 [Jatropha curcas] |
| 14 |
Hb_000428_030 |
0.0910871905 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 15 |
Hb_000665_260 |
0.091294443 |
- |
- |
PREDICTED: uncharacterized protein LOC105637620 isoform X1 [Jatropha curcas] |
| 16 |
Hb_004586_090 |
0.0939144546 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 16 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001133_090 |
0.0943511385 |
- |
- |
hypothetical protein POPTR_0012s02680g [Populus trichocarpa] |
| 18 |
Hb_000133_090 |
0.0947931196 |
- |
- |
PREDICTED: uncharacterized protein LOC105629320 [Jatropha curcas] |
| 19 |
Hb_159295_010 |
0.0950018145 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 20 |
Hb_003964_110 |
0.0962173717 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g26460, mitochondrial [Jatropha curcas] |