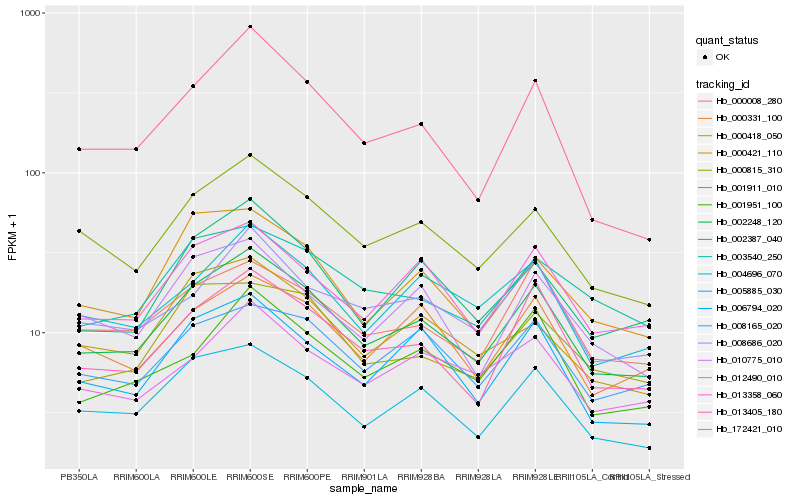
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002248_120 |
0.0 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 2 |
Hb_005885_030 |
0.0682093351 |
desease resistance |
Gene Name: TIR |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 3 |
Hb_001911_010 |
0.0719626195 |
- |
- |
PREDICTED: ALA-interacting subunit 3-like [Jatropha curcas] |
| 4 |
Hb_013405_180 |
0.0744274515 |
- |
- |
PREDICTED: uncharacterized protein LOC105648441 [Jatropha curcas] |
| 5 |
Hb_013358_060 |
0.0801545228 |
- |
- |
PREDICTED: uncharacterized protein LOC105637564 [Jatropha curcas] |
| 6 |
Hb_002387_040 |
0.0812860255 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 7 |
Hb_004696_070 |
0.0879540294 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like [Jatropha curcas] |
| 8 |
Hb_008686_020 |
0.0883667921 |
- |
- |
PREDICTED: uncharacterized protein LOC105632277 isoform X1 [Jatropha curcas] |
| 9 |
Hb_006794_020 |
0.0889733493 |
- |
- |
PREDICTED: uncharacterized protein LOC105645849 isoform X6 [Jatropha curcas] |
| 10 |
Hb_000331_100 |
0.090691497 |
- |
- |
PREDICTED: uncharacterized protein LOC105640211 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000008_280 |
0.0910227823 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 12 |
Hb_008165_020 |
0.09187121 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 10 isoform X1 [Jatropha curcas] |
| 13 |
Hb_172421_010 |
0.0921944665 |
- |
- |
cysteine protease [Hevea brasiliensis] |
| 14 |
Hb_001951_100 |
0.0926381438 |
- |
- |
Calcium-activated outward-rectifying potassium channel, putative [Ricinus communis] |
| 15 |
Hb_000418_050 |
0.0929849299 |
- |
- |
PREDICTED: uncharacterized protein LOC105636892 [Jatropha curcas] |
| 16 |
Hb_000421_110 |
0.0938647247 |
- |
- |
PREDICTED: V-type proton ATPase subunit a1 isoform X2 [Jatropha curcas] |
| 17 |
Hb_010775_010 |
0.0943470864 |
- |
- |
PREDICTED: uncharacterized protein LOC105642163 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000815_310 |
0.0953231304 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 19 |
Hb_012490_010 |
0.0953951077 |
- |
- |
PREDICTED: uncharacterized protein LOC105637916 isoform X2 [Jatropha curcas] |
| 20 |
Hb_003540_250 |
0.0969510141 |
- |
- |
PREDICTED: uncharacterized protein LOC105649465 isoform X2 [Jatropha curcas] |