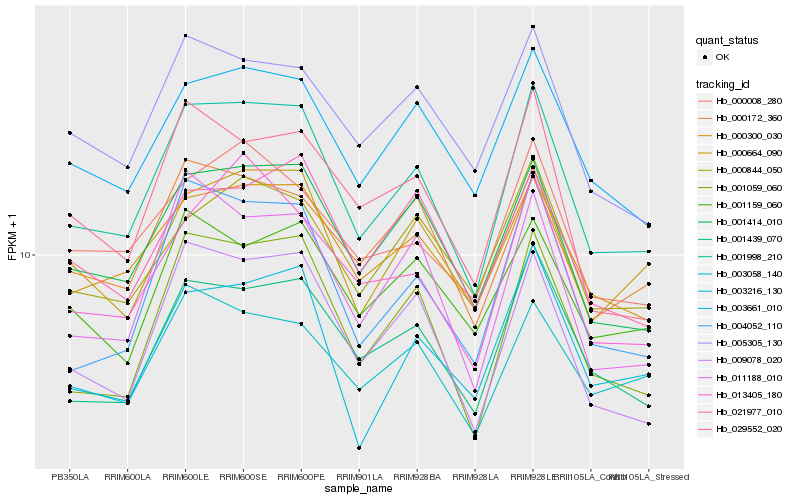
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001998_210 |
0.0 |
- |
- |
unnamed protein product [Coffea canephora] |
| 2 |
Hb_001414_010 |
0.069763565 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 3 |
Hb_001439_070 |
0.0714656175 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 4 |
Hb_001059_060 |
0.0775605951 |
- |
- |
PREDICTED: bifunctional riboflavin kinase/FMN phosphatase [Jatropha curcas] |
| 5 |
Hb_004052_110 |
0.0787501524 |
- |
- |
PREDICTED: host cell factor 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_009078_020 |
0.0792907936 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 7 |
Hb_000300_030 |
0.0800185965 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |
| 8 |
Hb_003661_010 |
0.0804420684 |
- |
- |
PREDICTED: zinc transporter 6, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000664_090 |
0.0851678437 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 10 |
Hb_011188_010 |
0.0885536403 |
- |
- |
PREDICTED: uncharacterized protein LOC105642145 isoform X1 [Jatropha curcas] |
| 11 |
Hb_005305_130 |
0.0905729216 |
- |
- |
PREDICTED: sulfite reductase [ferredoxin], chloroplastic [Jatropha curcas] |
| 12 |
Hb_003058_140 |
0.0910640849 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000172_360 |
0.0918720378 |
- |
- |
PREDICTED: apoptotic chromatin condensation inducer in the nucleus [Jatropha curcas] |
| 14 |
Hb_000008_280 |
0.0919819638 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 15 |
Hb_029552_020 |
0.0937096212 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000844_050 |
0.0951501904 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 17 |
Hb_021977_010 |
0.0964955533 |
- |
- |
PREDICTED: leukotriene A-4 hydrolase homolog [Jatropha curcas] |
| 18 |
Hb_003216_130 |
0.0970676506 |
- |
- |
PREDICTED: uncharacterized protein LOC105641500 [Jatropha curcas] |
| 19 |
Hb_013405_180 |
0.097133102 |
- |
- |
PREDICTED: uncharacterized protein LOC105648441 [Jatropha curcas] |
| 20 |
Hb_001159_060 |
0.0989016678 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |