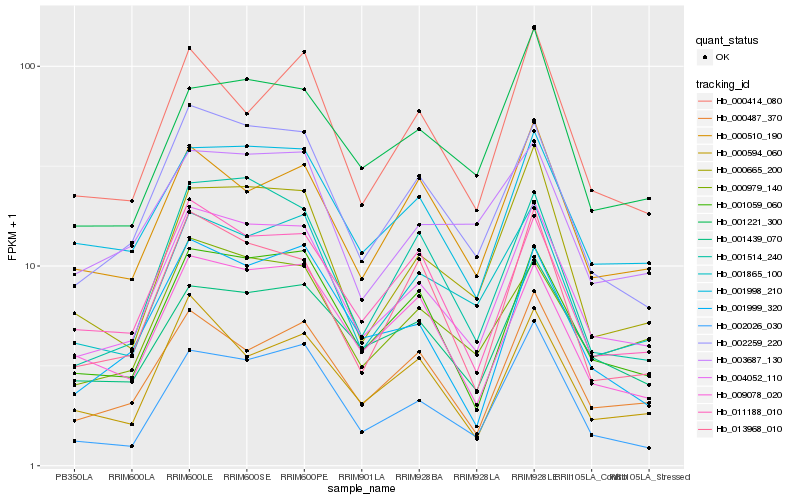
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004052_110 |
0.0 |
- |
- |
PREDICTED: host cell factor 2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002259_220 |
0.0665305523 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 66 [Jatropha curcas] |
| 3 |
Hb_001059_060 |
0.0727782259 |
- |
- |
PREDICTED: bifunctional riboflavin kinase/FMN phosphatase [Jatropha curcas] |
| 4 |
Hb_001998_210 |
0.0787501524 |
- |
- |
unnamed protein product [Coffea canephora] |
| 5 |
Hb_001865_100 |
0.0795663562 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9 [Jatropha curcas] |
| 6 |
Hb_000979_140 |
0.0805391629 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 7 |
Hb_011188_010 |
0.0885568857 |
- |
- |
PREDICTED: uncharacterized protein LOC105642145 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001439_070 |
0.0901652597 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 9 |
Hb_000487_370 |
0.0924850253 |
- |
- |
- |
| 10 |
Hb_000665_200 |
0.0936125074 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 11 |
Hb_009078_020 |
0.0950408298 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 12 |
Hb_001999_320 |
0.1003904384 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA [Jatropha curcas] |
| 13 |
Hb_001514_240 |
0.1008410465 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 14 |
Hb_013968_010 |
0.1022964523 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp3 [Jatropha curcas] |
| 15 |
Hb_000414_080 |
0.1024639124 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_003687_130 |
0.1036052371 |
- |
- |
PREDICTED: probable methyltransferase PMT3 [Jatropha curcas] |
| 17 |
Hb_002026_030 |
0.1038391757 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 18 |
Hb_000510_190 |
0.1053548569 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 19 |
Hb_000594_060 |
0.1077371957 |
- |
- |
PREDICTED: uncharacterized protein LOC105641988 [Jatropha curcas] |
| 20 |
Hb_001221_300 |
0.1082853947 |
- |
- |
PREDICTED: acetolactate synthase 3, chloroplastic [Jatropha curcas] |