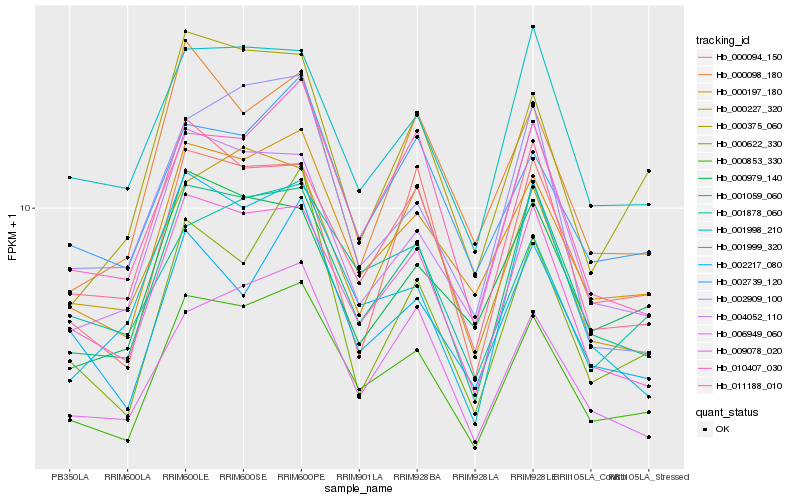
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000853_330 |
0.0 |
- |
- |
PREDICTED: importin subunit alpha-like [Jatropha curcas] |
| 2 |
Hb_001059_060 |
0.0716064086 |
- |
- |
PREDICTED: bifunctional riboflavin kinase/FMN phosphatase [Jatropha curcas] |
| 3 |
Hb_000197_180 |
0.0817305402 |
- |
- |
PREDICTED: bifunctional dihydrofolate reductase-thymidylate synthase 1 [Jatropha curcas] |
| 4 |
Hb_009078_020 |
0.0846885055 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 5 |
Hb_010407_030 |
0.1028086481 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 6 |
Hb_006949_060 |
0.1032471649 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 7 |
Hb_000375_060 |
0.1121016838 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1-like [Jatropha curcas] |
| 8 |
Hb_011188_010 |
0.1129832823 |
- |
- |
PREDICTED: uncharacterized protein LOC105642145 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001999_320 |
0.115873728 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor DIVARICATA [Jatropha curcas] |
| 10 |
Hb_000622_330 |
0.1160410158 |
- |
- |
PREDICTED: uncharacterized protein LOC105642906 [Jatropha curcas] |
| 11 |
Hb_001878_060 |
0.1179999061 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0007s15110g [Populus trichocarpa] |
| 12 |
Hb_002739_120 |
0.1189266935 |
- |
- |
PREDICTED: zinc finger protein ZPR1-like [Jatropha curcas] |
| 13 |
Hb_001998_210 |
0.1191503622 |
- |
- |
unnamed protein product [Coffea canephora] |
| 14 |
Hb_004052_110 |
0.1192178478 |
- |
- |
PREDICTED: host cell factor 2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000094_150 |
0.1219206892 |
- |
- |
- |
| 16 |
Hb_002217_080 |
0.1220428772 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 16 [Jatropha curcas] |
| 17 |
Hb_000227_320 |
0.1234176998 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_002909_100 |
0.1254901224 |
- |
- |
PREDICTED: uncharacterized protein LOC105648153 [Jatropha curcas] |
| 19 |
Hb_000098_180 |
0.1263471549 |
- |
- |
PREDICTED: uncharacterized protein LOC105633342 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000979_140 |
0.1276574031 |
- |
- |
kinase family protein [Populus trichocarpa] |