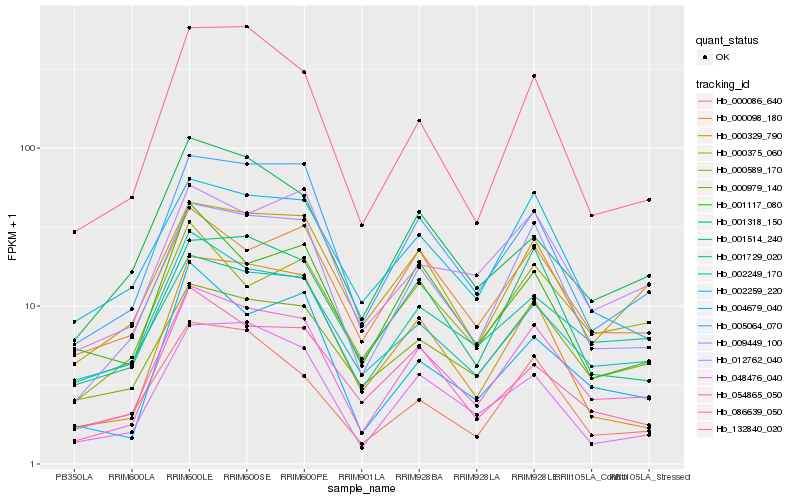
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_054865_050 |
0.0 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 2 |
Hb_009449_100 |
0.0732755001 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 1 [Jatropha curcas] |
| 3 |
Hb_005064_070 |
0.0940789923 |
- |
- |
PREDICTED: uncharacterized protein LOC105648318 [Jatropha curcas] |
| 4 |
Hb_002249_170 |
0.0985954631 |
- |
- |
copine, putative [Ricinus communis] |
| 5 |
Hb_000375_060 |
0.1012992138 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1-like [Jatropha curcas] |
| 6 |
Hb_001514_240 |
0.1064166404 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 7 |
Hb_000098_180 |
0.1069258075 |
- |
- |
PREDICTED: uncharacterized protein LOC105633342 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001729_020 |
0.1086877089 |
transcription factor |
TF Family: C3H |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_000979_140 |
0.1101848123 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 10 |
Hb_002259_220 |
0.1119845903 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 66 [Jatropha curcas] |
| 11 |
Hb_132840_020 |
0.1133568539 |
- |
- |
PREDICTED: protein LHY [Jatropha curcas] |
| 12 |
Hb_048476_040 |
0.1140666621 |
- |
- |
PREDICTED: uncharacterized protein LOC104611656 [Nelumbo nucifera] |
| 13 |
Hb_012762_040 |
0.1171716773 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |
| 14 |
Hb_001318_150 |
0.1225231847 |
- |
- |
PREDICTED: shaggy-related protein kinase alpha-like [Jatropha curcas] |
| 15 |
Hb_004679_040 |
0.1225268253 |
- |
- |
PREDICTED: rop guanine nucleotide exchange factor 1 [Jatropha curcas] |
| 16 |
Hb_086639_050 |
0.1229761697 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001117_080 |
0.1231331785 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000589_170 |
0.1250806253 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 19 |
Hb_000086_640 |
0.1255662833 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 20 |
Hb_000329_790 |
0.1260917144 |
- |
- |
PREDICTED: uncharacterized protein LOC105643367 [Jatropha curcas] |