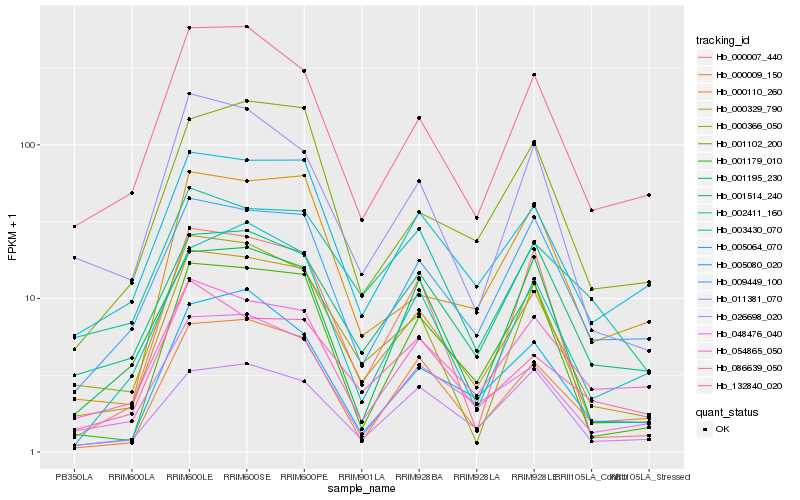
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000329_790 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643367 [Jatropha curcas] |
| 2 |
Hb_005064_070 |
0.0931877053 |
- |
- |
PREDICTED: uncharacterized protein LOC105648318 [Jatropha curcas] |
| 3 |
Hb_048476_040 |
0.1006155143 |
- |
- |
PREDICTED: uncharacterized protein LOC104611656 [Nelumbo nucifera] |
| 4 |
Hb_132840_020 |
0.1031688114 |
- |
- |
PREDICTED: protein LHY [Jatropha curcas] |
| 5 |
Hb_009449_100 |
0.1040268568 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 1 [Jatropha curcas] |
| 6 |
Hb_001179_010 |
0.1086044589 |
- |
- |
PREDICTED: glycosyltransferase family protein 64 protein C5 [Jatropha curcas] |
| 7 |
Hb_000366_050 |
0.111877621 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 8 |
Hb_001514_240 |
0.1120809893 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 9 |
Hb_011381_070 |
0.1130999272 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 10 |
Hb_000009_150 |
0.1150321985 |
- |
- |
carbohydrate transporter, putative [Ricinus communis] |
| 11 |
Hb_001102_200 |
0.1207747048 |
- |
- |
PREDICTED: trifunctional UDP-glucose 4,6-dehydratase/UDP-4-keto-6-deoxy-D-glucose 3,5-epimerase/UDP-4-keto-L-rhamnose-reductase RHM1 [Jatropha curcas] |
| 12 |
Hb_026698_020 |
0.1215874769 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 13 |
Hb_005080_020 |
0.1238400876 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 14 |
Hb_003430_070 |
0.1257011534 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |
| 15 |
Hb_054865_050 |
0.1260917144 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 16 |
Hb_000007_440 |
0.1269474134 |
- |
- |
PREDICTED: putative phospholipid-transporting ATPase 9 [Jatropha curcas] |
| 17 |
Hb_086639_050 |
0.1308135329 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 18 |
Hb_001195_230 |
0.1343513184 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 4 [Jatropha curcas] |
| 19 |
Hb_000110_260 |
0.1350024248 |
- |
- |
PREDICTED: uncharacterized protein LOC105642009 [Jatropha curcas] |
| 20 |
Hb_002411_160 |
0.1352997393 |
- |
- |
PREDICTED: uncharacterized protein LOC105631262 isoform X1 [Jatropha curcas] |