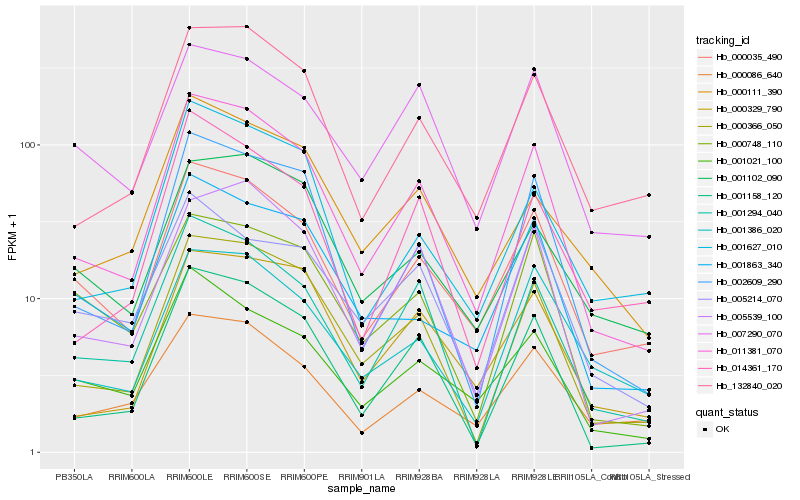
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011381_070 |
0.0 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 2 |
Hb_000366_050 |
0.0767692451 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 3 |
Hb_001158_120 |
0.0775808482 |
- |
- |
PREDICTED: uncharacterized protein LOC105648761 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000035_490 |
0.086757857 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 5 |
Hb_002609_290 |
0.0919609304 |
- |
- |
PREDICTED: non-specific phospholipase C1 [Jatropha curcas] |
| 6 |
Hb_001294_040 |
0.0940626037 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 7 |
Hb_132840_020 |
0.0986367846 |
- |
- |
PREDICTED: protein LHY [Jatropha curcas] |
| 8 |
Hb_005539_100 |
0.1011094664 |
- |
- |
nucleoside transporter, putative [Ricinus communis] |
| 9 |
Hb_000329_790 |
0.1130999272 |
- |
- |
PREDICTED: uncharacterized protein LOC105643367 [Jatropha curcas] |
| 10 |
Hb_001021_100 |
0.1161840985 |
- |
- |
PREDICTED: wee1-like protein kinase [Jatropha curcas] |
| 11 |
Hb_014361_170 |
0.1199147102 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 37 [Jatropha curcas] |
| 12 |
Hb_000086_640 |
0.124908972 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 13 |
Hb_001627_010 |
0.1250028011 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 14 |
Hb_000111_390 |
0.125400944 |
- |
- |
PREDICTED: uncharacterized protein LOC105631262 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001863_340 |
0.1290305719 |
- |
- |
PREDICTED: probable pectin methyltransferase QUA2 [Jatropha curcas] |
| 16 |
Hb_001386_020 |
0.1291759565 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_007290_070 |
0.1354753211 |
- |
- |
Monodehydroascorbate reductase family protein [Populus trichocarpa] |
| 18 |
Hb_001102_090 |
0.1366986034 |
- |
- |
PREDICTED: mitogen-activated protein kinase 19 [Jatropha curcas] |
| 19 |
Hb_005214_070 |
0.1378536915 |
- |
- |
PREDICTED: fatty acid amide hydrolase [Jatropha curcas] |
| 20 |
Hb_000748_110 |
0.1383465641 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |