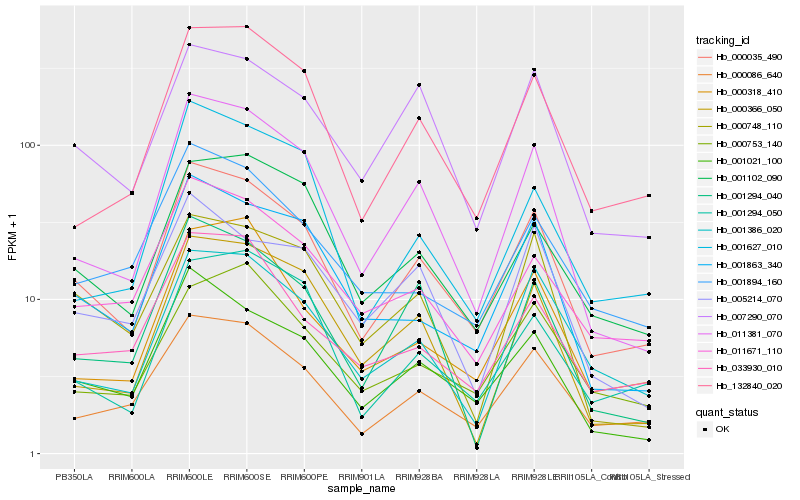
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000035_490 |
0.0 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 2 |
Hb_011381_070 |
0.086757857 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 3 |
Hb_000086_640 |
0.0887816394 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 4 |
Hb_001021_100 |
0.0930478221 |
- |
- |
PREDICTED: wee1-like protein kinase [Jatropha curcas] |
| 5 |
Hb_132840_020 |
0.108627819 |
- |
- |
PREDICTED: protein LHY [Jatropha curcas] |
| 6 |
Hb_001863_340 |
0.108981503 |
- |
- |
PREDICTED: probable pectin methyltransferase QUA2 [Jatropha curcas] |
| 7 |
Hb_011671_110 |
0.1142797406 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 8 |
Hb_007290_070 |
0.1160719239 |
- |
- |
Monodehydroascorbate reductase family protein [Populus trichocarpa] |
| 9 |
Hb_001102_090 |
0.1162971828 |
- |
- |
PREDICTED: mitogen-activated protein kinase 19 [Jatropha curcas] |
| 10 |
Hb_001386_020 |
0.118680357 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_033930_010 |
0.1214623375 |
- |
- |
PREDICTED: uncharacterized protein LOC105635519 [Jatropha curcas] |
| 12 |
Hb_001894_160 |
0.1284075994 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001294_050 |
0.1304148104 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPP13, putative [Ricinus communis] |
| 14 |
Hb_000318_410 |
0.1306550142 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1-like [Jatropha curcas] |
| 15 |
Hb_000753_140 |
0.1319976662 |
- |
- |
PREDICTED: uncharacterized protein LOC105640672 [Jatropha curcas] |
| 16 |
Hb_001294_040 |
0.1340879309 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 17 |
Hb_001627_010 |
0.1343020248 |
- |
- |
Alpha-1,4-glucan-protein synthase [UDP-forming], putative [Ricinus communis] |
| 18 |
Hb_005214_070 |
0.1354867819 |
- |
- |
PREDICTED: fatty acid amide hydrolase [Jatropha curcas] |
| 19 |
Hb_000748_110 |
0.1366002255 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |
| 20 |
Hb_000366_050 |
0.1372164646 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |