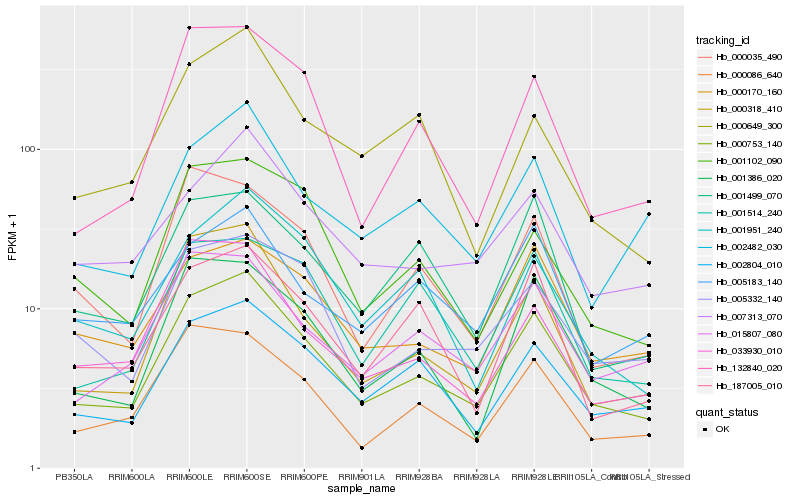
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000753_140 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640672 [Jatropha curcas] |
| 2 |
Hb_000318_410 |
0.0943971409 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1-like [Jatropha curcas] |
| 3 |
Hb_002804_010 |
0.1067189819 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like [Jatropha curcas] |
| 4 |
Hb_000086_640 |
0.1070502282 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 5 |
Hb_001386_020 |
0.108733638 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_015807_080 |
0.11532251 |
- |
- |
PREDICTED: helicase protein MOM1-like [Jatropha curcas] |
| 7 |
Hb_132840_020 |
0.1157706113 |
- |
- |
PREDICTED: protein LHY [Jatropha curcas] |
| 8 |
Hb_005332_140 |
0.1196587395 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000170_160 |
0.1225527822 |
- |
- |
PREDICTED: NAD kinase 2, chloroplastic [Jatropha curcas] |
| 10 |
Hb_007313_070 |
0.1236430769 |
- |
- |
PREDICTED: gibberellin receptor GID1C [Jatropha curcas] |
| 11 |
Hb_033930_010 |
0.1261729149 |
- |
- |
PREDICTED: uncharacterized protein LOC105635519 [Jatropha curcas] |
| 12 |
Hb_187005_010 |
0.1266438092 |
transcription factor |
TF Family: MYB-related |
PREDICTED: 1-phosphatidylinositol 3-phosphate 5-kinase [Jatropha curcas] |
| 13 |
Hb_002482_030 |
0.1277218459 |
transcription factor |
TF Family: PLATZ |
unknown [Medicago truncatula] |
| 14 |
Hb_001499_070 |
0.1299528862 |
- |
- |
PREDICTED: calcium-transporting ATPase 9, plasma membrane-type isoform X1 [Jatropha curcas] |
| 15 |
Hb_000035_490 |
0.1319976662 |
transcription factor |
TF Family: DBB |
PREDICTED: zinc finger protein CONSTANS-LIKE 4 [Jatropha curcas] |
| 16 |
Hb_001102_090 |
0.1325084159 |
- |
- |
PREDICTED: mitogen-activated protein kinase 19 [Jatropha curcas] |
| 17 |
Hb_001951_240 |
0.1330180591 |
- |
- |
PREDICTED: protein MEI2-like 5 [Jatropha curcas] |
| 18 |
Hb_001514_240 |
0.1336835706 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 19 |
Hb_005183_140 |
0.136524999 |
- |
- |
PREDICTED: protein WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 1-like [Jatropha curcas] |
| 20 |
Hb_000649_300 |
0.136997718 |
transcription factor |
TF Family: Tify |
JAZ9 [Hevea brasiliensis] |