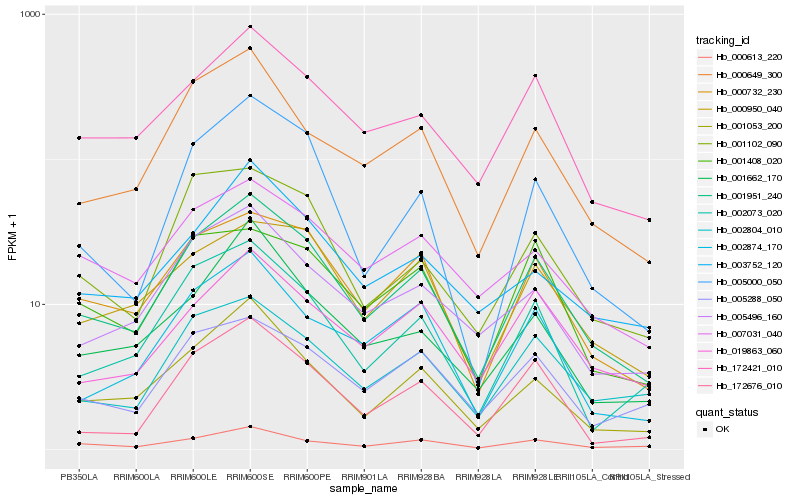
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001951_240 |
0.0 |
- |
- |
PREDICTED: protein MEI2-like 5 [Jatropha curcas] |
| 2 |
Hb_000732_230 |
0.0929975928 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_172421_010 |
0.0931064616 |
- |
- |
cysteine protease [Hevea brasiliensis] |
| 4 |
Hb_001053_200 |
0.0999133995 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 5 |
Hb_000649_300 |
0.1041347415 |
transcription factor |
TF Family: Tify |
JAZ9 [Hevea brasiliensis] |
| 6 |
Hb_002804_010 |
0.1069399949 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like [Jatropha curcas] |
| 7 |
Hb_002073_020 |
0.1118731353 |
- |
- |
PREDICTED: uncharacterized protein LOC105631164 [Jatropha curcas] |
| 8 |
Hb_005496_160 |
0.1144642625 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000613_220 |
0.1145572322 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 10 |
Hb_019863_060 |
0.1155806379 |
- |
- |
PREDICTED: target of Myb protein 1-like [Populus euphratica] |
| 11 |
Hb_000950_040 |
0.1159747449 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 12 |
Hb_005000_050 |
0.1178609414 |
- |
- |
PREDICTED: tetraspanin-2 [Jatropha curcas] |
| 13 |
Hb_001102_090 |
0.1195340796 |
- |
- |
PREDICTED: mitogen-activated protein kinase 19 [Jatropha curcas] |
| 14 |
Hb_002874_170 |
0.1199901498 |
- |
- |
PREDICTED: L-arabinokinase-like isoform X3 [Jatropha curcas] |
| 15 |
Hb_172676_010 |
0.1217317632 |
- |
- |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 16 |
Hb_005288_050 |
0.1222831541 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR4 [Jatropha curcas] |
| 17 |
Hb_003752_120 |
0.1254563494 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 18 |
Hb_007031_040 |
0.1260303678 |
transcription factor |
TF Family: bZIP |
PREDICTED: bZIP transcription factor 17-like [Jatropha curcas] |
| 19 |
Hb_001662_170 |
0.1266210689 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 20 |
Hb_001408_020 |
0.1288010632 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 2-like [Jatropha curcas] |