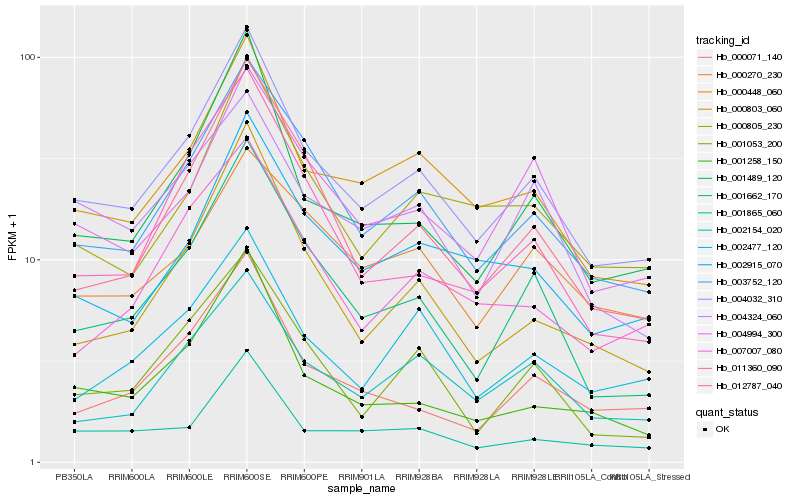
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004032_310 |
0.0 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 14 [Jatropha curcas] |
| 2 |
Hb_003752_120 |
0.0724352966 |
- |
- |
PREDICTED: probably inactive leucine-rich repeat receptor-like protein kinase At5g48380 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000448_060 |
0.0748659543 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 4 |
Hb_001489_120 |
0.0883846699 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001662_170 |
0.0898620128 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 6 |
Hb_004994_300 |
0.0997758071 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 7 |
Hb_002915_070 |
0.1005572698 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA GLABRA 1 [Jatropha curcas] |
| 8 |
Hb_001865_060 |
0.1009221756 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 9 |
Hb_002154_020 |
0.1032168424 |
- |
- |
protein with unknown function [Ricinus communis] |
| 10 |
Hb_000805_230 |
0.103918066 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 11 |
Hb_012787_040 |
0.1050164154 |
- |
- |
PREDICTED: uncharacterized protein LOC105649794 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000803_060 |
0.1086713462 |
- |
- |
poly(p)/ATP NAD kinase, putative [Ricinus communis] |
| 13 |
Hb_001053_200 |
0.1096338717 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 14 |
Hb_001258_150 |
0.1096762047 |
- |
- |
PREDICTED: COMPASS-like H3K4 histone methylase component WDR5A [Jatropha curcas] |
| 15 |
Hb_002477_120 |
0.1104554032 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 16 |
Hb_000071_140 |
0.1111115313 |
- |
- |
PREDICTED: uncharacterized protein LOC105628258 [Jatropha curcas] |
| 17 |
Hb_004324_060 |
0.1138243144 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 18 |
Hb_007007_080 |
0.1145013012 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_011360_090 |
0.1218572029 |
- |
- |
PREDICTED: uncharacterized protein LOC105644141 [Jatropha curcas] |
| 20 |
Hb_000270_230 |
0.1234511728 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 19 [Jatropha curcas] |