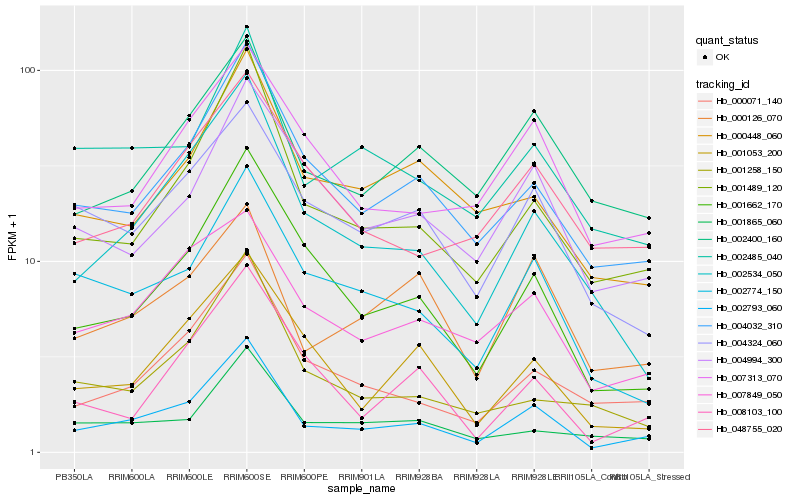
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002793_060 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g02980 [Populus euphratica] |
| 2 |
Hb_001489_120 |
0.1083147894 |
- |
- |
PREDICTED: alpha-amylase 3, chloroplastic [Jatropha curcas] |
| 3 |
Hb_004032_310 |
0.1251607916 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 14 [Jatropha curcas] |
| 4 |
Hb_001662_170 |
0.1300082024 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 5 |
Hb_002534_050 |
0.1326047758 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein JCGZ_11638 [Jatropha curcas] |
| 6 |
Hb_000071_140 |
0.1328823763 |
- |
- |
PREDICTED: uncharacterized protein LOC105628258 [Jatropha curcas] |
| 7 |
Hb_002774_150 |
0.1440785143 |
- |
- |
PREDICTED: F-box protein At1g22220 [Jatropha curcas] |
| 8 |
Hb_002485_040 |
0.1477550432 |
- |
- |
PREDICTED: uncharacterized protein LOC105637426 [Jatropha curcas] |
| 9 |
Hb_007849_050 |
0.1519271371 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 [Jatropha curcas] |
| 10 |
Hb_001053_200 |
0.1537814915 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 11 |
Hb_002400_160 |
0.1542228216 |
- |
- |
Ubiquitin-protein ligase BRE1A, putative [Ricinus communis] |
| 12 |
Hb_004324_060 |
0.1542270132 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 13 |
Hb_008103_100 |
0.1569957864 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPS2 [Jatropha curcas] |
| 14 |
Hb_007313_070 |
0.157095788 |
- |
- |
PREDICTED: gibberellin receptor GID1C [Jatropha curcas] |
| 15 |
Hb_000126_070 |
0.1584393383 |
- |
- |
putative leucine-rich repeat receptor-like serine/threonine-protein kinase [Gossypium arboreum] |
| 16 |
Hb_004994_300 |
0.1588129288 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 17 |
Hb_000448_060 |
0.1597957032 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 2 [Jatropha curcas] |
| 18 |
Hb_001865_060 |
0.1600332881 |
- |
- |
PREDICTED: uncharacterized protein LOC105647962 [Jatropha curcas] |
| 19 |
Hb_048755_020 |
0.1646496045 |
- |
- |
hypothetical protein JCGZ_09860 [Jatropha curcas] |
| 20 |
Hb_001258_150 |
0.1653194831 |
- |
- |
PREDICTED: COMPASS-like H3K4 histone methylase component WDR5A [Jatropha curcas] |