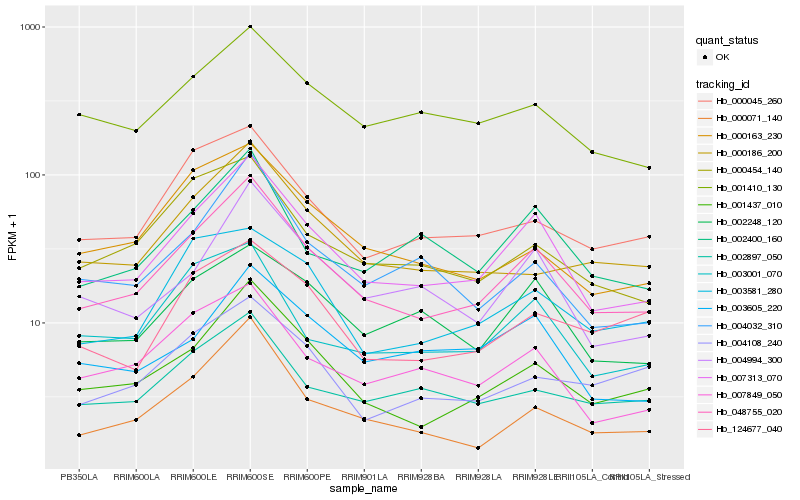
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_048755_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_09860 [Jatropha curcas] |
| 2 |
Hb_007313_070 |
0.0461885126 |
- |
- |
PREDICTED: gibberellin receptor GID1C [Jatropha curcas] |
| 3 |
Hb_001437_010 |
0.0783852993 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g33260 [Jatropha curcas] |
| 4 |
Hb_000071_140 |
0.1072756153 |
- |
- |
PREDICTED: uncharacterized protein LOC105628258 [Jatropha curcas] |
| 5 |
Hb_000045_260 |
0.1147937882 |
- |
- |
protein with unknown function [Ricinus communis] |
| 6 |
Hb_004994_300 |
0.1158479714 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 7 |
Hb_000186_200 |
0.1191055946 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 8 |
Hb_007849_050 |
0.1195879698 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 [Jatropha curcas] |
| 9 |
Hb_000454_140 |
0.1197704192 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2a-like [Jatropha curcas] |
| 10 |
Hb_002400_160 |
0.1203921034 |
- |
- |
Ubiquitin-protein ligase BRE1A, putative [Ricinus communis] |
| 11 |
Hb_001410_130 |
0.1205236276 |
- |
- |
hypothetical protein POPTR_0001s10880g [Populus trichocarpa] |
| 12 |
Hb_000163_230 |
0.1206527238 |
- |
- |
Phytochrome A-associated F-box protein, putative [Ricinus communis] |
| 13 |
Hb_002897_050 |
0.1223126709 |
- |
- |
PREDICTED: uncharacterized protein LOC105649158 [Jatropha curcas] |
| 14 |
Hb_003001_070 |
0.1231489197 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0002s24750g [Populus trichocarpa] |
| 15 |
Hb_004032_310 |
0.1249236565 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 14 [Jatropha curcas] |
| 16 |
Hb_003581_280 |
0.1281539547 |
- |
- |
KINASE 2B family protein [Populus trichocarpa] |
| 17 |
Hb_002248_120 |
0.1282868024 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 18 |
Hb_003605_220 |
0.1300699453 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 19 |
Hb_124677_040 |
0.1304546564 |
- |
- |
PREDICTED: uncharacterized protein LOC105650471 [Jatropha curcas] |
| 20 |
Hb_004108_240 |
0.1310808161 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |