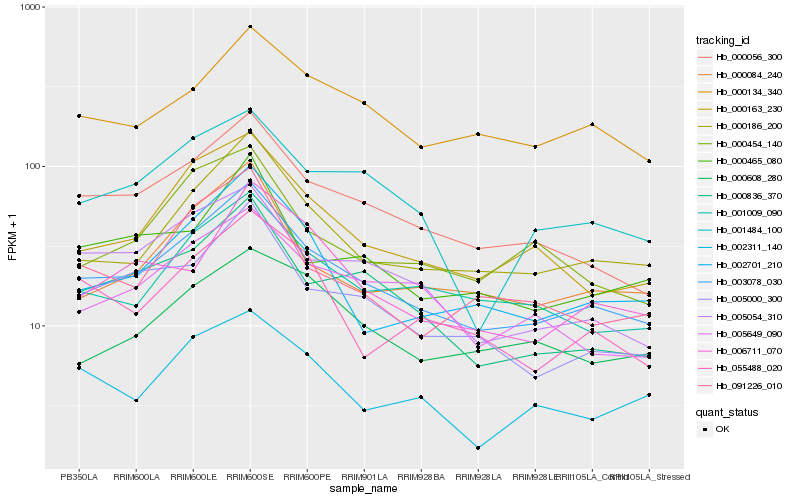
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003078_030 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CHIP [Jatropha curcas] |
| 2 |
Hb_000056_300 |
0.0726084421 |
- |
- |
PREDICTED: probable methyltransferase PMT2 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000186_200 |
0.0820868098 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 4 |
Hb_000163_230 |
0.0915377703 |
- |
- |
Phytochrome A-associated F-box protein, putative [Ricinus communis] |
| 5 |
Hb_001009_090 |
0.0923844458 |
- |
- |
CDK protein [Hevea brasiliensis] |
| 6 |
Hb_005000_300 |
0.0953181873 |
transcription factor |
TF Family: TRAF |
putative regulatory protein NPR1 [Hevea brasiliensis] |
| 7 |
Hb_000836_370 |
0.0958352855 |
- |
- |
PREDICTED: protein REVEILLE 3-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000134_340 |
0.095997528 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 5 [Jatropha curcas] |
| 9 |
Hb_005054_310 |
0.0983678212 |
- |
- |
PREDICTED: glycosyltransferase family 64 protein C4 [Jatropha curcas] |
| 10 |
Hb_000084_240 |
0.1029541223 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g57790-like [Jatropha curcas] |
| 11 |
Hb_002701_210 |
0.103241223 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 12 |
Hb_001484_100 |
0.1062627481 |
- |
- |
PREDICTED: cyanate hydratase [Jatropha curcas] |
| 13 |
Hb_005649_090 |
0.1066133975 |
- |
- |
PREDICTED: ninja-family protein AFP3-like [Jatropha curcas] |
| 14 |
Hb_000465_080 |
0.1076056566 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RBBP6-like [Jatropha curcas] |
| 15 |
Hb_091226_010 |
0.1105242909 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA isoform X1 [Jatropha curcas] |
| 16 |
Hb_055488_020 |
0.1146066338 |
- |
- |
PREDICTED: uncharacterized protein LOC105641270 [Jatropha curcas] |
| 17 |
Hb_002311_140 |
0.11540357 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 15-like [Jatropha curcas] |
| 18 |
Hb_000454_140 |
0.1155939809 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2a-like [Jatropha curcas] |
| 19 |
Hb_006711_070 |
0.1162273289 |
- |
- |
hypothetical protein RCOM_1336320 [Ricinus communis] |
| 20 |
Hb_000608_280 |
0.1190502304 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |