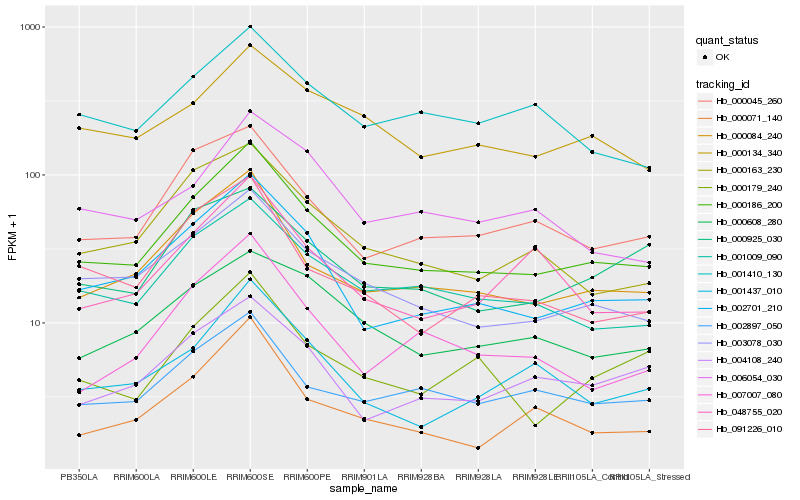
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000186_200 |
0.0 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 2 |
Hb_002701_210 |
0.0723956235 |
- |
- |
Protein COBRA precursor, putative [Ricinus communis] |
| 3 |
Hb_000084_240 |
0.0756619752 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g57790-like [Jatropha curcas] |
| 4 |
Hb_003078_030 |
0.0820868098 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CHIP [Jatropha curcas] |
| 5 |
Hb_001009_090 |
0.0916472602 |
- |
- |
CDK protein [Hevea brasiliensis] |
| 6 |
Hb_000045_260 |
0.0970323982 |
- |
- |
protein with unknown function [Ricinus communis] |
| 7 |
Hb_000163_230 |
0.100910462 |
- |
- |
Phytochrome A-associated F-box protein, putative [Ricinus communis] |
| 8 |
Hb_002897_050 |
0.1014985549 |
- |
- |
PREDICTED: uncharacterized protein LOC105649158 [Jatropha curcas] |
| 9 |
Hb_091226_010 |
0.1116397517 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA isoform X1 [Jatropha curcas] |
| 10 |
Hb_004108_240 |
0.1125606761 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 11 |
Hb_001437_010 |
0.1150436277 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g33260 [Jatropha curcas] |
| 12 |
Hb_007007_080 |
0.1162281148 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000134_340 |
0.1165751446 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 5 [Jatropha curcas] |
| 14 |
Hb_000608_280 |
0.1182825014 |
- |
- |
Aspartic proteinase Asp1 precursor, putative [Ricinus communis] |
| 15 |
Hb_048755_020 |
0.1191055946 |
- |
- |
hypothetical protein JCGZ_09860 [Jatropha curcas] |
| 16 |
Hb_000179_240 |
0.1206130474 |
- |
- |
PREDICTED: threonine synthase 1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001410_130 |
0.1227880454 |
- |
- |
hypothetical protein POPTR_0001s10880g [Populus trichocarpa] |
| 18 |
Hb_006054_030 |
0.1228171252 |
- |
- |
PREDICTED: protein GPR107-like [Jatropha curcas] |
| 19 |
Hb_000925_030 |
0.1232735047 |
- |
- |
ATP-dependent RNA helicase, putative [Ricinus communis] |
| 20 |
Hb_000071_140 |
0.1237753453 |
- |
- |
PREDICTED: uncharacterized protein LOC105628258 [Jatropha curcas] |