| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
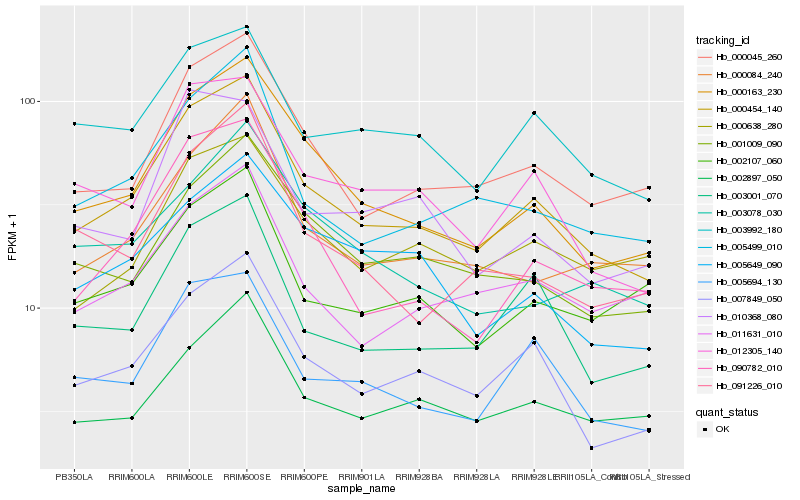
Hb_000454_140 |
0.0 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2a-like [Jatropha curcas] |
| 2 |
Hb_000163_230 |
0.0672475215 |
- |
- |
Phytochrome A-associated F-box protein, putative [Ricinus communis] |
| 3 |
Hb_007849_050 |
0.0750113397 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 [Jatropha curcas] |
| 4 |
Hb_000045_260 |
0.0804831973 |
- |
- |
protein with unknown function [Ricinus communis] |
| 5 |
Hb_012305_140 |
0.0875557533 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 5-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_005694_130 |
0.0879850025 |
- |
- |
PREDICTED: U-box domain-containing protein 44-like [Jatropha curcas] |
| 7 |
Hb_005499_010 |
0.0910433017 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |
| 8 |
Hb_003001_070 |
0.0910863477 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0002s24750g [Populus trichocarpa] |
| 9 |
Hb_003992_180 |
0.0957304925 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11 isoform X1 [Jatropha curcas] |
| 10 |
Hb_090782_010 |
0.09640034 |
- |
- |
PREDICTED: diacylglycerol kinase 5-like [Jatropha curcas] |
| 11 |
Hb_000084_240 |
0.0986564643 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g57790-like [Jatropha curcas] |
| 12 |
Hb_002897_050 |
0.1028097119 |
- |
- |
PREDICTED: uncharacterized protein LOC105649158 [Jatropha curcas] |
| 13 |
Hb_001009_090 |
0.1035856847 |
- |
- |
CDK protein [Hevea brasiliensis] |
| 14 |
Hb_011631_010 |
0.1061541517 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 15 |
Hb_000638_280 |
0.107245182 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 16 |
Hb_002107_060 |
0.1078580658 |
- |
- |
PREDICTED: uncharacterized protein LOC105635312 [Jatropha curcas] |
| 17 |
Hb_091226_010 |
0.1087406909 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA isoform X1 [Jatropha curcas] |
| 18 |
Hb_005649_090 |
0.1134977662 |
- |
- |
PREDICTED: ninja-family protein AFP3-like [Jatropha curcas] |
| 19 |
Hb_003078_030 |
0.1155939809 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase CHIP [Jatropha curcas] |
| 20 |
Hb_010368_080 |
0.1165529564 |
- |
- |
PREDICTED: uncharacterized protein LOC105630287 [Jatropha curcas] |