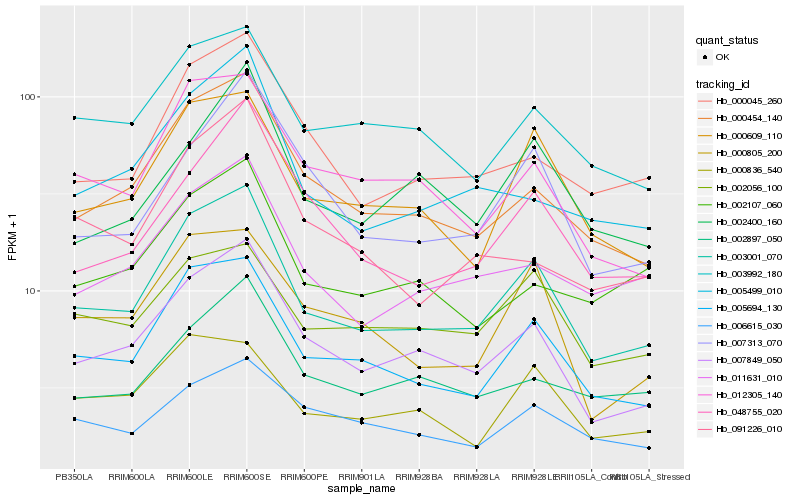
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003001_070 |
0.0 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0002s24750g [Populus trichocarpa] |
| 2 |
Hb_005694_130 |
0.0621655192 |
- |
- |
PREDICTED: U-box domain-containing protein 44-like [Jatropha curcas] |
| 3 |
Hb_007849_050 |
0.0776588446 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 [Jatropha curcas] |
| 4 |
Hb_000454_140 |
0.0910863477 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2a-like [Jatropha curcas] |
| 5 |
Hb_000609_110 |
0.0933372773 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11-like [Jatropha curcas] |
| 6 |
Hb_012305_140 |
0.1062096127 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 5-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_003992_180 |
0.1079752835 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000045_260 |
0.1089085446 |
- |
- |
protein with unknown function [Ricinus communis] |
| 9 |
Hb_011631_010 |
0.1104989231 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 10 |
Hb_000805_200 |
0.1113894449 |
- |
- |
PREDICTED: telomere repeat-binding protein 2 isoform X2 [Jatropha curcas] |
| 11 |
Hb_005499_010 |
0.1156472353 |
transcription factor |
TF Family: TRAF |
Regulatory protein NPR1, putative [Ricinus communis] |
| 12 |
Hb_002056_100 |
0.1194672172 |
transcription factor |
TF Family: C2H2 |
PREDICTED: probable lysine-specific demethylase ELF6 [Jatropha curcas] |
| 13 |
Hb_000836_540 |
0.1195612192 |
- |
- |
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein [Theobroma cacao] |
| 14 |
Hb_007313_070 |
0.1223554293 |
- |
- |
PREDICTED: gibberellin receptor GID1C [Jatropha curcas] |
| 15 |
Hb_002897_050 |
0.1226193344 |
- |
- |
PREDICTED: uncharacterized protein LOC105649158 [Jatropha curcas] |
| 16 |
Hb_048755_020 |
0.1231489197 |
- |
- |
hypothetical protein JCGZ_09860 [Jatropha curcas] |
| 17 |
Hb_006615_030 |
0.1260535596 |
- |
- |
prolyl oligopeptidase family protein [Populus trichocarpa] |
| 18 |
Hb_002107_060 |
0.127429972 |
- |
- |
PREDICTED: uncharacterized protein LOC105635312 [Jatropha curcas] |
| 19 |
Hb_002400_160 |
0.1283936244 |
- |
- |
Ubiquitin-protein ligase BRE1A, putative [Ricinus communis] |
| 20 |
Hb_091226_010 |
0.1318910549 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA isoform X1 [Jatropha curcas] |