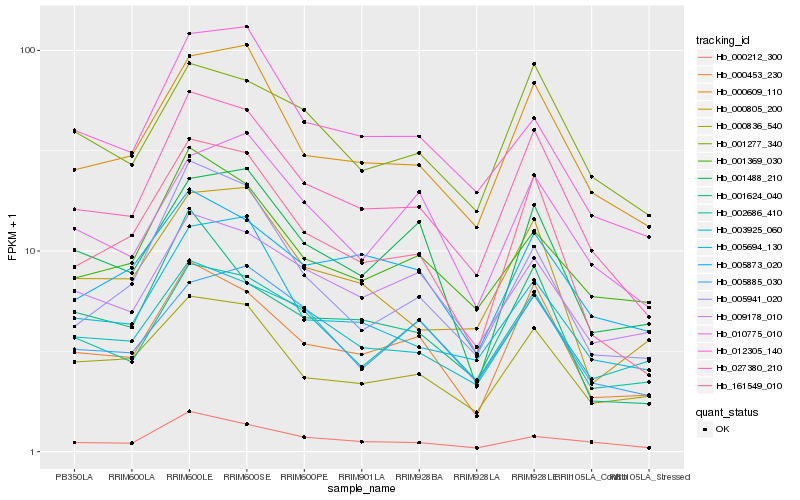
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027380_210 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634030 [Jatropha curcas] |
| 2 |
Hb_161549_010 |
0.0715974726 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000609_110 |
0.0716547928 |
- |
- |
PREDICTED: methyl-CpG-binding domain-containing protein 11-like [Jatropha curcas] |
| 4 |
Hb_000453_230 |
0.0810525292 |
- |
- |
PREDICTED: NO-associated protein 1, chloroplastic/mitochondrial [Jatropha curcas] |
| 5 |
Hb_005694_130 |
0.0991927224 |
- |
- |
PREDICTED: U-box domain-containing protein 44-like [Jatropha curcas] |
| 6 |
Hb_000836_540 |
0.0999809903 |
- |
- |
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein [Theobroma cacao] |
| 7 |
Hb_000212_300 |
0.1015010972 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 8 |
Hb_002686_410 |
0.1028038886 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 48 kDa protein-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_001277_340 |
0.1082176322 |
- |
- |
aldo/keto reductase, putative [Ricinus communis] |
| 10 |
Hb_001369_030 |
0.1087542711 |
- |
- |
PREDICTED: ceramide kinase isoform X1 [Jatropha curcas] |
| 11 |
Hb_000805_200 |
0.1091052027 |
- |
- |
PREDICTED: telomere repeat-binding protein 2 isoform X2 [Jatropha curcas] |
| 12 |
Hb_012305_140 |
0.110099902 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 5-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_003925_060 |
0.1126056973 |
- |
- |
PREDICTED: uncharacterized protein LOC105645218 isoform X1 [Jatropha curcas] |
| 14 |
Hb_005873_020 |
0.1138018553 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 15 |
Hb_005885_030 |
0.1160990702 |
desease resistance |
Gene Name: TIR |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 16 |
Hb_010775_010 |
0.1174319016 |
- |
- |
PREDICTED: uncharacterized protein LOC105642163 isoform X1 [Jatropha curcas] |
| 17 |
Hb_005941_020 |
0.1180157574 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |
| 18 |
Hb_009178_010 |
0.1190061112 |
- |
- |
PREDICTED: uncharacterized protein LOC105637474 [Jatropha curcas] |
| 19 |
Hb_001624_040 |
0.119936971 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 5 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001488_210 |
0.1209615261 |
- |
- |
PREDICTED: protein RIK [Jatropha curcas] |