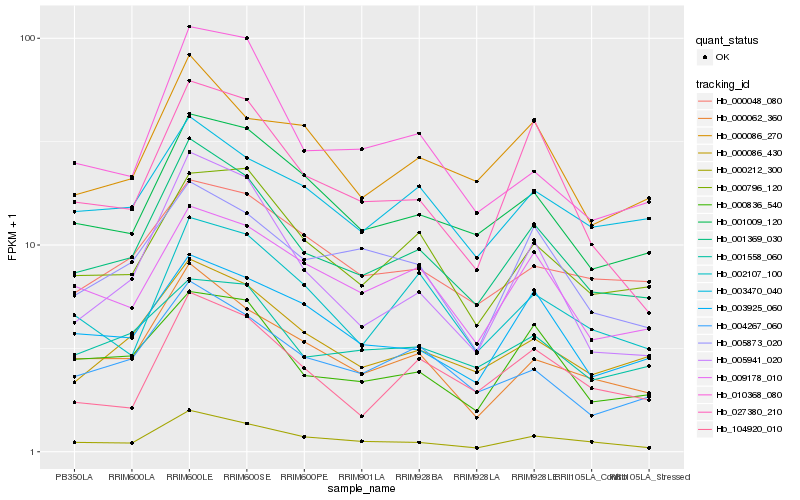
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001369_030 |
0.0 |
- |
- |
PREDICTED: ceramide kinase isoform X1 [Jatropha curcas] |
| 2 |
Hb_000086_430 |
0.0813468021 |
- |
- |
hypothetical protein JCGZ_17649 [Jatropha curcas] |
| 3 |
Hb_000212_300 |
0.0836259936 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 4 |
Hb_000062_360 |
0.0841108972 |
- |
- |
PREDICTED: DNA polymerase alpha subunit B [Jatropha curcas] |
| 5 |
Hb_001009_120 |
0.0888545156 |
- |
- |
PREDICTED: transcription factor GTE10 isoform X2 [Jatropha curcas] |
| 6 |
Hb_004267_060 |
0.0915340168 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_000836_540 |
0.094716131 |
- |
- |
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein [Theobroma cacao] |
| 8 |
Hb_000796_120 |
0.0951737875 |
- |
- |
PREDICTED: SWR1-complex protein 4 isoform X3 [Jatropha curcas] |
| 9 |
Hb_001558_060 |
0.1000592383 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 17 isoform X2 [Jatropha curcas] |
| 10 |
Hb_009178_010 |
0.101446751 |
- |
- |
PREDICTED: uncharacterized protein LOC105637474 [Jatropha curcas] |
| 11 |
Hb_000086_270 |
0.1058518802 |
- |
- |
hypothetical protein POPTR_0002s03730g [Populus trichocarpa] |
| 12 |
Hb_003925_060 |
0.1059601766 |
- |
- |
PREDICTED: uncharacterized protein LOC105645218 isoform X1 [Jatropha curcas] |
| 13 |
Hb_005873_020 |
0.1076264282 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 14 |
Hb_027380_210 |
0.1087542711 |
- |
- |
PREDICTED: uncharacterized protein LOC105634030 [Jatropha curcas] |
| 15 |
Hb_002107_100 |
0.109056017 |
- |
- |
PREDICTED: la-related protein 1C-like isoform X3 [Jatropha curcas] |
| 16 |
Hb_003470_040 |
0.1091519431 |
- |
- |
PREDICTED: 2-oxoisovalerate dehydrogenase subunit beta 1, mitochondrial [Jatropha curcas] |
| 17 |
Hb_010368_080 |
0.1091575436 |
- |
- |
PREDICTED: uncharacterized protein LOC105630287 [Jatropha curcas] |
| 18 |
Hb_000048_080 |
0.1092217531 |
- |
- |
PREDICTED: cyclin-dependent kinase D-3 isoform X1 [Jatropha curcas] |
| 19 |
Hb_104920_010 |
0.1098955128 |
- |
- |
hypothetical protein VITISV_031859 [Vitis vinifera] |
| 20 |
Hb_005941_020 |
0.1101035743 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 2 [Jatropha curcas] |