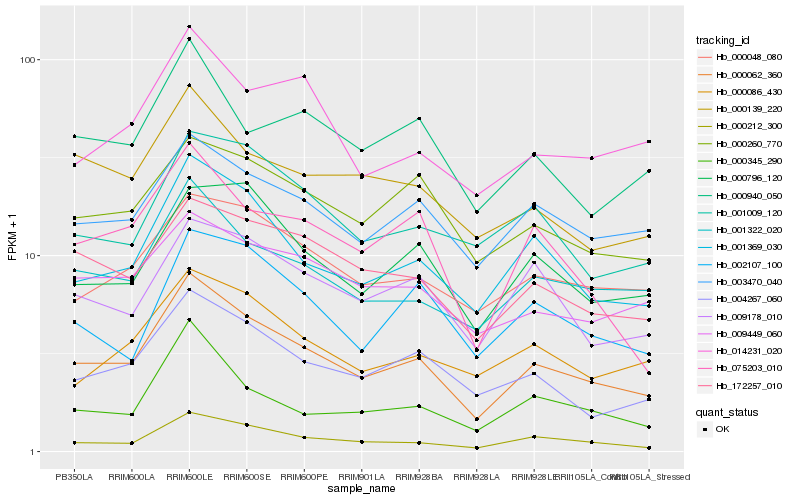
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000062_360 |
0.0 |
- |
- |
PREDICTED: DNA polymerase alpha subunit B [Jatropha curcas] |
| 2 |
Hb_001369_030 |
0.0841108972 |
- |
- |
PREDICTED: ceramide kinase isoform X1 [Jatropha curcas] |
| 3 |
Hb_000212_300 |
0.0885733754 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 4 |
Hb_001322_020 |
0.0959761512 |
- |
- |
PREDICTED: polycomb group protein VERNALIZATION 2-like isoform X2 [Populus euphratica] |
| 5 |
Hb_004267_060 |
0.1010776687 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 6 |
Hb_172257_010 |
0.1080419494 |
transcription factor |
TF Family: HMG |
high mobility group family protein [Populus trichocarpa] |
| 7 |
Hb_003470_040 |
0.1091986442 |
- |
- |
PREDICTED: 2-oxoisovalerate dehydrogenase subunit beta 1, mitochondrial [Jatropha curcas] |
| 8 |
Hb_000796_120 |
0.1141699094 |
- |
- |
PREDICTED: SWR1-complex protein 4 isoform X3 [Jatropha curcas] |
| 9 |
Hb_000048_080 |
0.1158914896 |
- |
- |
PREDICTED: cyclin-dependent kinase D-3 isoform X1 [Jatropha curcas] |
| 10 |
Hb_075203_010 |
0.1161143643 |
- |
- |
PREDICTED: calcium-dependent protein kinase SK5 isoform X1 [Jatropha curcas] |
| 11 |
Hb_009178_010 |
0.1163216841 |
- |
- |
PREDICTED: uncharacterized protein LOC105637474 [Jatropha curcas] |
| 12 |
Hb_000086_430 |
0.1169466673 |
- |
- |
hypothetical protein JCGZ_17649 [Jatropha curcas] |
| 13 |
Hb_000139_220 |
0.1172285571 |
- |
- |
leucine rich repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_000260_770 |
0.1175357872 |
- |
- |
er lumen protein retaining receptor, putative [Ricinus communis] |
| 15 |
Hb_000345_290 |
0.1192289614 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_009449_060 |
0.1198168671 |
- |
- |
PREDICTED: ubiquitin-like-specific protease 1D isoform X2 [Jatropha curcas] |
| 17 |
Hb_001009_120 |
0.1198983001 |
- |
- |
PREDICTED: transcription factor GTE10 isoform X2 [Jatropha curcas] |
| 18 |
Hb_002107_100 |
0.119986478 |
- |
- |
PREDICTED: la-related protein 1C-like isoform X3 [Jatropha curcas] |
| 19 |
Hb_000940_050 |
0.1207581503 |
- |
- |
hypothetical protein JCGZ_19588 [Jatropha curcas] |
| 20 |
Hb_014231_020 |
0.1214150039 |
- |
- |
PREDICTED: ras-related protein Rab11D [Jatropha curcas] |